Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Roger D. Jones | -- | 2122 | 2024-02-21 01:14:57 | | | |
| 2 | Fanny Huang | -20 word(s) | 2102 | 2024-02-21 06:19:44 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Jones, R.D. Information Transmission in G Protein-Coupled Receptors. Encyclopedia. Available online: https://encyclopedia.pub/entry/55255 (accessed on 28 February 2026).
Jones RD. Information Transmission in G Protein-Coupled Receptors. Encyclopedia. Available at: https://encyclopedia.pub/entry/55255. Accessed February 28, 2026.
Jones, Roger D.. "Information Transmission in G Protein-Coupled Receptors" Encyclopedia, https://encyclopedia.pub/entry/55255 (accessed February 28, 2026).
Jones, R.D. (2024, February 21). Information Transmission in G Protein-Coupled Receptors. In Encyclopedia. https://encyclopedia.pub/entry/55255
Jones, Roger D.. "Information Transmission in G Protein-Coupled Receptors." Encyclopedia. Web. 21 February, 2024.
Copy Citation
G protein-coupled receptors (GPCRs) are the largest class of receptors in the human genome and constitute about 30% of all drug targets. The concept of information capacity can be used to measure researcher's understanding of GPCR computation. Capacity is the maximum amount of information that can be transmitted by a system.
G protein-coupled receptors
information transmission
1. Introduction
Most papers on G protein-coupled receptors (GPCRs) start out with some variation of the sentence, “GPCRs are the largest class of receptors in the human genome and constitute about 30% of all drug targets [1]”. The foreseeable practical applications of this class of proteins are well established. The long-term practical and scientific applications may be even more interesting, however. GPCRs are molecular microprocessors, or perhaps more accurately “nanoprocessors,” that transmit, process, and compare information among and about the environments on each side of the cell membrane and within the membrane itself [2][3]. GPCRs are important examples of molecular computation that may shed light on the mechanisms of information processing in all adaptable biological processes. If that is true, then it may be scientifically and practically profitable to regard GPCRs within the context of general information theory and statistical physics, two disciplines with connections to broad sets of general principles.
Figure 1 is a schematic of the GPCR complex. The core of the complex is composed of seven transmembrane (7TM) alpha helices that form a barrel. There is an eighth intracellular helix that plays an important role in information transmission. A ligand in the extracellular region can bind to a pocket in the barrel and allosterically affect the conformation of the intracellular portion of the core. The intracellular portion of the complex is composed of a collection of molecular switches that can take two forms: (1) a 𝐺𝛼 switch that involves the 𝐺𝛼 subunit of a G protein and (2) a collection of phosphorylation sites can form phosphorylation dephosphorylation cycles [4]. The phosphorylation sites form a barcode [5][6] that transmits information to 𝛽 arrestin (𝛽arr), which directs downstream responses to the ligand.
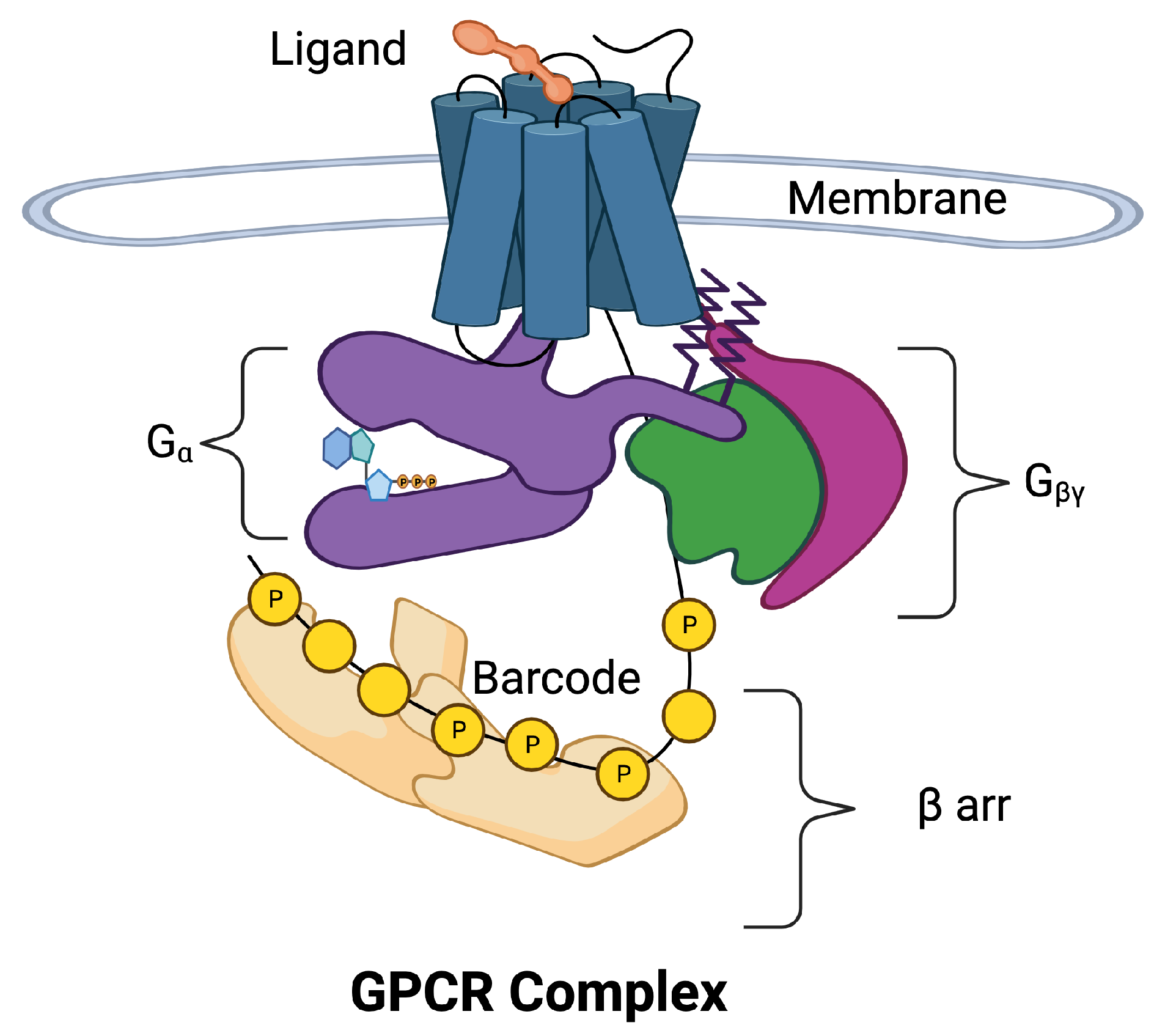
Figure 1. The 7-transmembrane GPCR is illustrated with blue cylinders representing the seven 𝛼 helices that span the cell membrane. The extracellular ligand (orange) binds to the binding site of the GPCR inducing movement in the 𝛼 helices. The helices allosterically alter the conformation of the intracellular domains of the GPCR complex. The intracellular portion of the complex has been separated for visibility. Two pathways may be activated, the 𝐺𝛼 pathway (purple) and the 𝛽arr pathway (tan). The 𝐺𝛼 subunit is a part of the G protein also composed of subunits 𝛽 (green) and 𝛾 (magenta). The 𝛽arr pathway is composed of additional response pathways determined by phosphorylation sites on the C tail of the GPCR and intracellular loops that form a barcode that encodes signals for downstream processes.
Even at this basic level, the picture of the GPCR complex is mysterious and unsatisfying. Intuitively, researchers see that a small amount of information is associated with the binding of a ligand to the GPCR, and perhaps the concentration of the ligand is translated into a larger amount of information that requires a barcode to store and transmit all the response options. Precision/personalized/stratified medicine attempts to identify the individual prognosis and targeted treatment at the right time for the right patient, or at least for smaller and more homogeneous groups [7][8][9]. The nuances of the information transmission across the membrane must be clear.
The study of information transmission is taking place on three scales. The largest scale studies take place at the level of the human organism, the clinical level (e.g., [10][11]). Here, approved drug treatments are applied to patient populations and advanced statistical tools are used to tease out the individual patient characteristics that respond differentially to specific treatments. This is a coarse approach to precision drug discovery.
At the smallest scales are detailed observations of GPCR structure (e.g., [6][12]). Here, portions of the GPCR complex are studied in isolation. This may involve, for instance, isolation of the C tail of the GPCR [6] or isolation of the GPCR core [12]. Often, synthetic nanobodies are used to mimic missing parts of the complex. Function and interaction among the complex of components is inferred from the structural observations.
Assay experiments (e.g., [13][14]) take place on intermediate scales. Here, GPCR complexes that are intact, or at least chimeric, are treated with ligands and the downstream responses are observed. Precision drug targets in the complex must be inferred.
Proper characterization of information transmission in the control of downstream response requires that the observations on all three scales be glued together into a single coherent picture. If physics is taken as an example, this glue is provided by theory that is able to fill in gaps in the picture. Recently a theory has been developed, the BOIS Model [15], that may be a candidate for filling in some of the unobserved gaps in the multi-scale observations. The model combines standard principles of statistical mechanics [16] with speculations on natural selection to make a series of predictions, some of which are testable and some of which lie in the gaps of the unobservable.
2. Observations of Information Transmission across the Cell Membrane
2.1. Direct Measurement of Information Transmission in GPCRs
Information capacity can be measured from assay experiments. However, an important challenge to the measurement of information flow in these experiments is removing signal from noise. Accurate measurements require the application of multiple ligand concentrations to a single cell [14][17]. Ref. [14] focused on the muscarinic acetylcholine receptor (M3R). The muscarinic receptor-induced calcium response measured in individual HEK293 cells was repeatedly stimulated with the ligand acetylcholine. Using this approach, single cell assays in human embryonic kidney 293 (HEK293) cells found a capacity greater than two bits of information [14][18].
These results are in contrast with some previous studies using cell populations that provided lower values for the information capacity of the GPCR pathways [19]. Lower capacity measurements were a consequence of increased noise due to variable responses among cells and the fact that individual cells were only exposed to a single value of ligand concentration [14][17].
2.2. Observations of Information Transmission in Assay Experiments
Bias between two downstream response pathways was examined in Ref. [13]. The authors focused on two receptors, the adrenergic receptor 𝛽2𝐴𝑅 and the angiotensin II receptor 𝐴𝑇1𝐴𝑅. Eleven different ligands were applied to 𝛽2𝐴𝑅, whereas ten were applied to 𝐴𝑇1𝐴𝑅. The measured downstream responses were 𝛽-arrestin (𝛽arr) recruitment to the GPCR as well as cAMP for 𝛽2𝐴𝑅 and 𝐼𝑃1 for 𝐴𝑇1𝐴𝑅 for the 𝐺𝛼 pathway. To reduce noise in 𝛽arr response, the C terminus of the human 𝛽2𝐴𝑅 was replaced with the C-terminal tail of the V2 vasopressin receptor tail. In addition to the bias observations this study identified various values of half maximal effective concentration (𝐸𝐶50) for the responses. The 𝐸𝐶50 for each response can be used to identify the ligand concentration at which the response is activated. This is in agreement with Ref. [14] where information about the ligand concentration was identified in the information transmission.
2.3. Observations of Allosteric Mechanisms of Information Transmission
The two signaling pathways, 𝐺𝛼 and 𝛽arr mediate distinct physiological effects [12][20]. In the prototypical angiotensin II receptor, for instance, 𝐺𝛼 coupling increases blood pressure, whereas 𝛽arr coupling promotes heart protection [21][22][23]. In opioid receptors, the 𝐺𝛼 pathway confers pain relief whereas the 𝛽arr path may be associated with side effects such as tolerance, dependence, addiction, constipation, and respiratory depression [24][25][26][27][28].
The GPCR protein is composed of seven alpha helices arranged in a barrel conformation and one intracellular helix (Figure 2A). The helices themselves are somewhat rigid whereas the connections between the helices are flexible. The determination of which parts of the protein are flexible and which are rigid is determined by the ratio of the local bond strength to the background energy fluctuations [29][30][31][32][33][34]. At thermal equilibrium at room temperature, the background energy fluctuations are at about 0.5 kcal/mol. The amount of energy available in a single ATP is about 12 kcal/mol ([35], [Sec. 15.2]). Typical covalent bond energies are around 80 kcal/mol and greater, whereas ionic, hydrogen, and hydrophobic interaction energies are typically around 5 kcal/mol ([36], Chapter 8). Van der Waals interactions are typically 0.5–1 kcal/mol [35]. Parts of the protein with bond energy less than the fluctuation energy are unstable leading to flexibility. Parts of the protein where individual bonds, or collections of bonds, that have bonding energies greater than the fluctuation energy are rigid. The flexible and rigid parts of a protein are not fixed. If the background fluctuation level changes energy, then flexible parts of the protein can become rigid or rigid parts of the protein can become flexible. Recent observations have discovered a major hydrogen-bond network that bathes the ligand and transmits information allosterically [12].
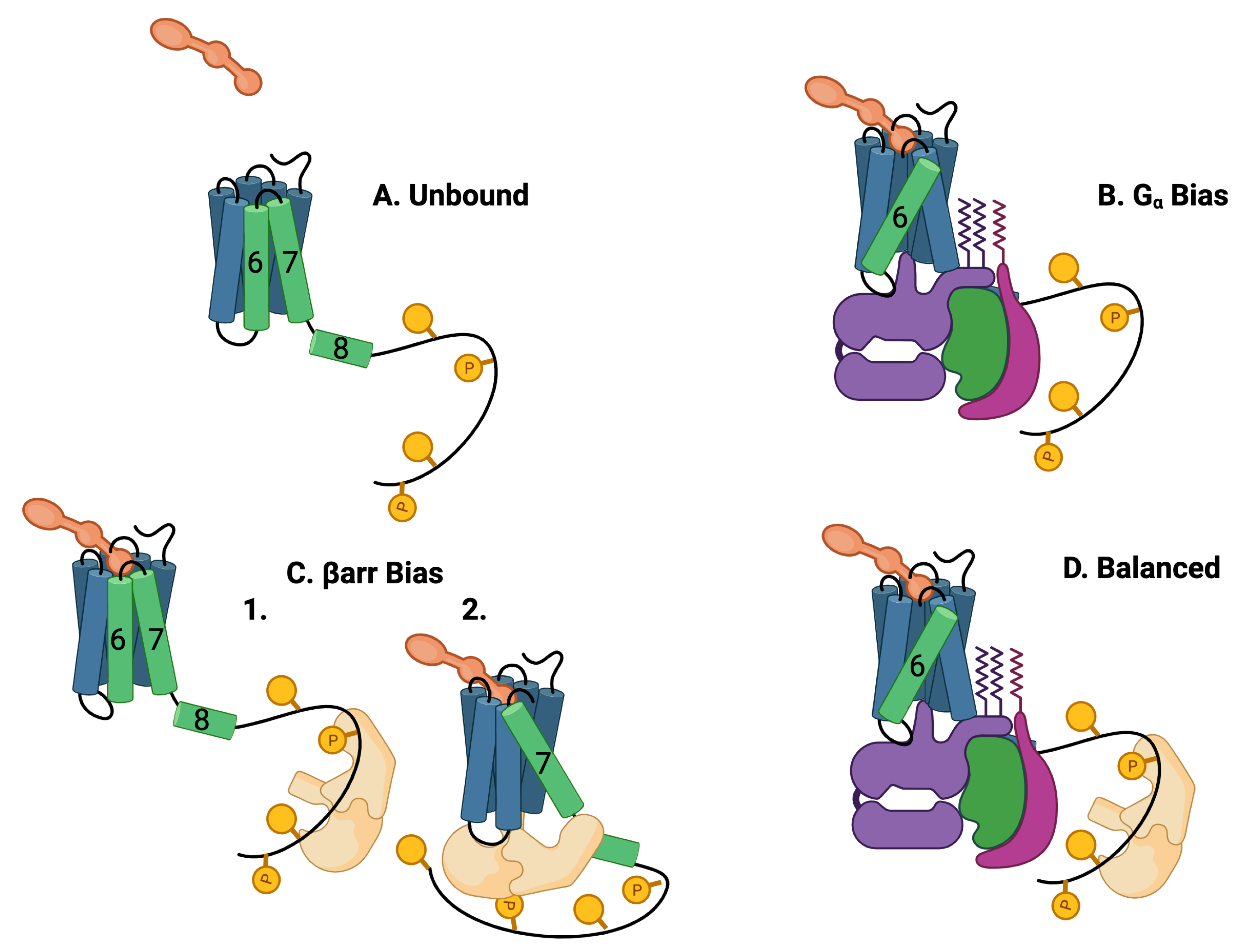
Figure 2. Five theoretically possible GPCR complex configurations, assuming that 𝐺𝛼 and 𝛽arr cannot occupy the intracellular core pocket simultaneously. (A) Unbound: The GPCR is composed of seven transmembrane (7TM) alpha helices and one intracellular helix labeled (8). The alpha helices are rigid and are connected by flexible amino-acid chains. The C tail of the GPCR may be quite long and contain multiple phosphorylation sites. Particularly important helices are in green. (B) 𝑮𝜶 bias: The ligand (orange) and the G protein (purple, green, and violet) may bind to the 7TM in a ternary reaction in which both the ligand and the G protein alter the 7 TM conformation. The most common conformational indicator is the rotation and extension of helix 6 (green). This configuration is G biased. (C) 𝜷arr bias: 𝜷arr may bind to the C tail of the GPCR (1) or 𝜷arr may also replace the G protein in the intracellular binding pocket (2). The conformation is characterized by a rotation and extension of helix 7. This conformation blocks the G-protein downstream response pathways. The C tail may also bind to the 𝜷arr that may select 𝜷arr downstream responses. This conformation is 𝜷arr biased. (D) Balanced: Another possibility is that the G protein and the 𝜷arr are both bound to the 7TM, the G protein bound to the 7TM pocket, and the 𝜷arr bound to the C-tail phosphorylation sites.
Another part of the GPCR, the C tail, is very flexible at room temperature and can take on many different conformations ([37][38][39]). The flexible/rigid nature of the complex conformations indicates that the complex can occupy multiple active quasi-stable states (Figure 2) [40].
Typically, binding of 𝐺𝛼 and 𝛽arr to the intracellular pocket of the GPCR is mediated by helices 6, 7, 8, and 5 [41] and the intracellular loops [42]. The 7TM conformation is altered from its baseline configuration (Figure 2A) when a ligand and a G protein form a ternary bond (Figure 2B) with the transmembrane portion of the GPCR [20]. This altered conformation transmits information from the extracellular environment to the intracellular environment. The G protein responds by activating downstream responses. The typical allosteric signature of this event is the rotation and extension of helix 6 [43]. Since only downstream responses triggered by the G protein are activated, this event is known as 𝐺𝛼 bias after the relevant 𝛼 subunit of the G protein. It has recently been found from mutation studies that receptor sites 𝑁1113.35 𝐴 and 𝑁2947.45 𝐴 induce biased signaling to 𝐺𝛼 and 𝛽arr, respectively, [12]. AngII is a balanced agonist that activates both 𝐺𝛼 and 𝛽-arrestin signaling pathways. AngII can be modified to generate AT1R-biased agonists, which could preferentially activate either signaling pathway [21][44][45].
The binding of the beta arrestin (𝛽arr) to the GPCR is observed to have more conformations than G-protein binding [6] as well as having more interactions with the membrane environment [46]. The 𝛽arr interacts simultaneously with the core GPCR in a ternary reaction [47][48][49]. Two distinct binding processes for 𝛽arr have been identified, dubbed “core-engaged” (Figure 2(C2)) and “tail-engaged” (Figure 2(C1)) for recruitment to the core and tail, respectively, [40][48]. This is illustrated in Figure 2C,D where a G protein occupies the core pocket in Figure 2D that is occupied by 𝛽arr in Figure 2(C2). The conformation of the 𝛽arr is different in the two binding paths [50]. Interestingly, 𝛽arr may continue to trigger downstream responses even after dissociation from the GPCR [39][51][52]. A typical signature for the 𝛽arr core-binding (Figure 2C) is the rotation and extension of helices 7 and 8 [43].
2.4. Information Transmitted to the Barcode
The barcode structure indicates that much more information may be stored in the barcode than the approximately two bits of information transmitted to the barcode [3][5][6][12][38][53][54][55][56][57]. As suggested in Ref. [14], this extra information may be about the details of the ligand concentration. This information may not be observed in the experiments [12][13][14] because of the focus on only the 𝐺𝛼 pathway and a single 𝛽arr recruitment pathway. In many cases, in these experiments, chimeric receptors were used that completely replaced the C tail of the receptor with a foreign C tail [13]. Except for the recruitment site, the phosphorylation sites on the foreign tail need not correspond with sites on the native tail. The use of the foreign tail removes information-storage capacity in the tail. The number of downstream pathways observable from a chimeric receptor would be two.
References
- Santos, R.; Ursu, O.; Gaulton, A.; Bento, A.P.; Donadi, R.S.; Bologa, C.G.; Karlsson, A.; Al-Lazikani, B.; Hersey, A.; Oprea, T.I.; et al. A comprehensive map of molecular drug targets. Nat. Rev. Drug Discov. 2017, 16, 19–34.
- Smith, J.S.; Lefkowitz, R.J.; Rajagopal, S. Biased signalling: From simple switches to allosteric microprocessors. Nat. Rev. Drug Discov. 2018, 17, 243–260.
- Chen, H.; Zhang, S.; Zhang, X.; Liu, H. QR code model: A new possibility for GPCR phosphorylation recognition. Cell Commun. Signal. 2022, 20, 23.
- Qian, H.; Reluga, T.C. Nonequilibrium thermodynamics and nonlinear kinetics in a cellular signaling switch. Phys. Rev. Lett. 2005, 94, 028101.
- Liu, Y.H.; Smith, S.; Mihalas, S.; Shea-Brown, E.; Sümbül, U. Cell-type–specific neuromodulation guides synaptic credit assignment in a spiking neural network. Proc. Natl. Acad. Sci. USA 2021, 118, e2111821118.
- Latorraca, N.R.; Masureel, M.; Hollingsworth, S.A.; Heydenreich, F.M.; Suomivuori, C.M.; Brinton, C.; Townshend, R.J.; Bouvier, M.; Kobilka, B.K.; Dror, R.O. How GPCR phosphorylation patterns orchestrate arrestin-mediated signaling. Cell 2020, 183, 1813–1825.
- Koenig, I.R.; Fuchs, O.; Hansen, G.; von Mutius, E.; Kopp, M.V. What is precision medicine? Eur. Respir. J. 2017, 50, 1700391.
- Kosorok, M.R.; Laber, E.B. Precision medicine. Annu. Rev. Stat. Its Appl. 2019, 6, 263–286.
- Ginsburg, G.S.; Phillips, K.A. Precision medicine: From science to value. Health Aff. 2018, 37, 694–701.
- Eder, S.; Leierer, J.; Kerschbaum, J.; Rosivall, L.; Wiecek, A.; de Zeeuw, D.; Mark, P.B.; Heinze, G.; Rossing, P.; Heerspink, H.L.; et al. A prospective cohort study in patients with type 2 diabetes mellitus for validation of biomarkers (PROVALID)—Study design and baseline characteristics. Kidney Blood Press. Res. 2018, 43, 181–190.
- Jones, R.D.; Abebe, S.; Distefano, V.; Mayer, G.; Poli, I.; Silvestri, C.; Slanzi, D. Candidate composite biomarker to inform drug treatments for diabetic kidney disease. Front. Med. 2023, 10, 1271407.
- Zhang, D.; Liu, Y.; Zaidi, S.A.; Xu, L.; Zhan, Y.; Chen, A.; Guo, J.; Huang, X.P.; Roth, B.L.; Katritch, V.; et al. Structural insights into angiotensin receptor signaling modulation by balanced and biased agonists. EMBO J. 2023, 42, e112940.
- Rajagopal, S.; Ahn, S.; Rominger, D.H.; Gowen-MacDonald, W.; Lam, C.M.; DeWire, S.M.; Violin, J.D.; Lefkowitz, R.J. Quantifying ligand bias at seven-transmembrane receptors. Mol. Pharmacol. 2011, 80, 367–377.
- Keshelava, A.; Solis, G.P.; Hersch, M.; Koval, A.; Kryuchkov, M.; Bergmann, S.; Katanaev, V.L. High capacity in G protein-coupled receptor signaling. Nat. Commun. 2018, 9, 876.
- Jones, R.D.; Jones, A.M. Model of Ligand-Triggered Information Transmission in G-Protein Coupled Receptor Complexes. Front. Endocrinol. 2023, 14, 879.
- Reif, F. Fundamentals of Statistical and Thermal Physics; Waveland Press: Long Grove, IL, USA, 2009.
- Zielińska, K.; Katanaev, V. Information theory: New look at oncogenic signaling pathways. Trends Cell Biol. 2019, 29, 862–875.
- Selimkhanov, J.; Taylor, B.; Yao, J.; Pilko, A.; Albeck, J.; Hoffmann, A.; Tsimring, L.; Wollman, R. Accurate information transmission through dynamic biochemical signaling networks. Science 2014, 346, 1370–1373.
- Cheong, R.; Rhee, A.; Wang, C.J.; Nemenman, I.; Levchenko, A. Information transduction capacity of noisy biochemical signaling networks. Science 2011, 334, 354–358.
- Lefkowitz, R. Nobel Lecture: A Brief History of G Protein Coupled Receptors. Nobel Lect. 2012, 52, 6366–6378.
- Violin, J.D.; DeWire, S.M.; Yamashita, D.; Rominger, D.H.; Nguyen, L.; Schiller, K.; Whalen, E.J.; Gowen, M.; Lark, M.W. Selectively engaging β-arrestins at the angiotensin II type 1 receptor reduces blood pressure and increases cardiac performance. J. Pharmacol. Exp. Ther. 2010, 335, 572–579.
- Felker, G.M.; Butler, J.; Collins, S.P.; Cotter, G.; Davison, B.A.; Ezekowitz, J.A.; Filippatos, G.; Levy, P.D.; Metra, M.; Ponikowski, P.; et al. Heart failure therapeutics on the basis of a biased ligand of the angiotensin-2 type 1 receptor: Rationale and design of the BLAST-AHF study (Biased Ligand of the Angiotensin Receptor Study in Acute Heart Failure). JACC Heart Fail. 2015, 3, 193–201.
- Takezako, T.; Unal, H.; Karnik, S.S.; Node, K. Current topics in angiotensin II type 1 receptor research: Focus on inverse agonism, receptor dimerization and biased agonism. Pharmacol. Res. 2017, 123, 40–50.
- Faouzi, A.; Varga, B.R.; Majumdar, S. Biased opioid ligands. Molecules 2020, 25, 4257.
- Che, T.; Dwivedi-Agnihotri, H.; Shukla, A.K.; Roth, B.L. Biased ligands at opioid receptors: Current status and future directions. Sci. Signal. 2021, 14, eaav0320.
- Slosky, L.M.; Caron, M.G.; Barak, L.S. Biased allosteric modulators: New frontiers in GPCR drug discovery. Trends Pharmacol. Sci. 2021, 42, 283–299.
- Hales, T. Arresting the development of morphine tolerance and dependence. Br. J. Anaesth. 2011, 107, 653–655.
- Bohn, L.M.; Gainetdinov, R.R.; Lin, F.T.; Lefkowitz, R.J.; Caron, M.G. μ-Opioid receptor desensitization by β-arrestin-2 determines morphine tolerance but not dependence. Nature 2000, 408, 720–723.
- Punjani, A.; Fleet, D.J. 3DFlex: Determining structure and motion of flexible proteins from cryo-EM. Nat. Methods 2023, 20, 860–870.
- Strokach, A.; Becerra, D.; Corbi-Verge, C.; Perez-Riba, A.; Kim, P.M. Fast and flexible protein design using deep graph neural networks. Cell Syst. 2020, 11, 402–411.
- Chiu, J.; Hogg, P.J. Allosteric disulfides: Sophisticated molecular structures enabling flexible protein regulation. J. Biol. Chem. 2019, 294, 2949–5908.
- Santos, K.B.; Guedes, I.A.; Karl, A.L.; Dardenne, L.E. Highly flexible ligand docking: Benchmarking of the DockThor program on the LEADS-PEP protein–peptide data set. J. Chem. Inf. Model. 2020, 60, 667–683.
- Zhou, Y.; Millott, R.; Kim, H.J.; Peng, S.; Edwards, R.A.; Skene-Arnold, T.; Hammel, M.; Lees-Miller, S.P.; Tainer, J.A.; Holmes, C.F.; et al. Flexible tethering of ASPP proteins facilitates PP-1c catalysis. Structure 2019, 27, 1485–1496.
- Michalak, D. Characterization of Disordered Biomolecular Systems with Scattering Techniques: A Flexible Protein Complex and Solid-Supported Lipid Bilayers. Ph.D. Thesis, Carnegie Mellon University, Pittsburgh, PA, USA, 2021.
- Berg Jeremy, M.; Tymoczko John, L.; Gatto, G.J., Jr.; Lubert, S. Biochemistry; W. H. Freeman: New York, NY, USA, 2019.
- CCCOnline. Anatomy & Physiology. Available online: https://pressbooks.ccconline.org/bio106/ (accessed on 14 December 2023).
- Kahsai, A.W.; Pani, B.; Lefkowitz, R.J. GPCR signaling: Conformational activation of arrestins. Cell Res. 2018, 28, 783–784.
- Latorraca, N.R.; Wang, J.K.; Bauer, B.; Townshend, R.J.; Hollingsworth, S.A.; Olivieri, J.E.; Xu, H.E.; Sommer, M.E.; Dror, R.O. Molecular mechanism of GPCR-mediated arrestin activation. Nature 2018, 557, 452–456.
- Eichel, K.; Jullié, D.; Barsi-Rhyne, B.; Latorraca, N.R.; Masureel, M.; Sibarita, J.B.; Dror, R.O.; von Zastrow, M. Catalytic activation of β-arrestin by GPCRs. Nature 2018, 557, 381–386.
- Gurevich, V.V.; Gurevich, E.V. Biased GPCR signaling: Possible mechanisms and inherent limitations. Pharmacol. Ther. 2020, 211, 107540.
- Hilger, D. The role of structural dynamics in GPCR-mediated signaling. FEBS J. 2021, 288, 2461–2489.
- Wang, Q.; Zhang, S.; Han, Z.; Fan, H.; Li, C. An investigation into the allosteric mechanism of GPCR A2A adenosine receptor with trajectory-based information theory and complex network model. J. Biomol. Struct. Dyn. 2021, 39, 6431–6439.
- Seyedabadi, M.; Ghahremani, M.H.; Albert, P.R. Biased signaling of G protein coupled receptors (GPCRs): Molecular determinants of GPCR/transducer selectivity and therapeutic potential. Pharmacol. Ther. 2019, 200, 148–178.
- Holloway, A.C.; Qian, H.; Pipolo, L.; Ziogas, J.; Miura, S.i.; Karnik, S.; Southwell, B.R.; Lew, M.J.; Thomas, W.G. Side-chain substitutions within angiotensin II reveal different requirements for signaling, internalization, and phosphorylation of type 1A angiotensin receptors. Mol. Pharmacol. 2002, 61, 768–777.
- Strachan, R.T.; Sun, J.p.; Rominger, D.H.; Violin, J.D.; Ahn, S.; Thomsen, A.R.B.; Zhu, X.; Kleist, A.; Costa, T.; Lefkowitz, R.J. Divergent transducer-specific molecular efficacies generate biased agonism at a G protein-coupled receptor (GPCR). J. Biol. Chem. 2014, 289, 14211–14224.
- Grimes, J.; Koszegi, Z.; Lanoiselée, Y.; Miljus, T.; O’Brien, S.L.; Stepniewski, T.M.; Medel-Lacruz, B.; Baidya, M.; Makarova, M.; Mistry, R.; et al. Plasma membrane preassociation drives β-arrestin coupling to receptors and activation. Cell 2023, 186, 2238–2255.
- Ma, N.; Nivedha, A.K.; Vaidehi, N. Allosteric communication regulates ligand-specific GPCR activity. FEBS J. 2021, 288, 2502–2512.
- Maharana, J.; Banerjee, R.; Yadav, M.K.; Sarma, P.; Shukla, A.K. Emerging structural insights into GPCR–β-arrestin interaction and functional outcomes. Curr. Opin. Struct. Biol. 2022, 75, 102406.
- Dwivedi-Agnihotri, H.; Chaturvedi, M.; Baidya, M.; Stepniewski, T.M.; Pandey, S.; Maharana, J.; Srivastava, A.; Caengprasath, N.; Hanyaloglu, A.C.; Selent, J.; et al. Distinct phosphorylation sites in a prototypical GPCR differently orchestrate β-arrestin interaction, trafficking, and signaling. Sci. Adv. 2020, 6, eabb8368.
- Shiraishi, Y.; Kofuku, Y.; Ueda, T.; Pandey, S.; Dwivedi-Agnihotri, H.; Shukla, A.K.; Shimada, I. Biphasic activation of β-arrestin 1 upon interaction with a GPCR revealed by methyl-TROSY NMR. Nat. Commun. 2021, 12, 7158.
- Eichel, K.; Jullié, D.; Von Zastrow, M. β-Arrestin drives MAP kinase signalling from clathrin-coated structures after GPCR dissociation. Nat. Cell Biol. 2016, 18, 303–310.
- Ranjan, R.; Gupta, P.; Shukla, A.K. GPCR Signaling: β-arrestins Kiss and Remember. Curr. Biol. 2016, 26, R285–R288.
- Tobin, A.B.; Butcher, A.J.; Kong, K.C. Location, location, location...site-specific GPCR phosphorylation offers a mechanism for cell-type-specific signalling. Trends Pharmacol. Sci. 2008, 29, 413–420.
- Tobin, A. G-protein-coupled receptor phosphorylation: Where, when and by whom. Br. J. Pharmacol. 2008, 153, S167–S176.
- Yang, Z.; Yang, F.; Zhang, D.; Liu, Z.; Lin, A.; Liu, C.; Xiao, P.; Yu, X.; Sun, J.P. Phosphorylation of G protein-coupled receptors: From the barcode hypothesis to the flute model. Mol. Pharmacol. 2017, 92, 201–210.
- Chakravorty, D.; Assmann, S.M. G protein subunit phosphorylation as a regulatory mechanism in heterotrimeric G protein signaling in mammals, yeast, and plants. Biochem. J. 2018, 475, 3331–3357.
- Tunc-Ozdemir, M.; Li, B.; Jaiswal, D.K.; Urano, D.; Jones, A.M.; Torres, M.P. Predicted functional implications of phosphorylation of regulator of G protein signaling protein in plants. Front. Plant Sci. 2017, 8, 1456.
More
Information
Subjects:
Cell & Tissue Engineering
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
565
Revisions:
2 times
(View History)
Update Date:
21 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





