Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bo Zhou | -- | 2845 | 2024-02-15 11:56:31 | | | |
| 2 | Lindsay Dong | Meta information modification | 2845 | 2024-02-18 01:56:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zhou, B.; Zheng, B.; Wu, W. Environmental Stress-Induced Anthocyanin Accumulation in Plants. Encyclopedia. Available online: https://encyclopedia.pub/entry/55069 (accessed on 08 February 2026).
Zhou B, Zheng B, Wu W. Environmental Stress-Induced Anthocyanin Accumulation in Plants. Encyclopedia. Available at: https://encyclopedia.pub/entry/55069. Accessed February 08, 2026.
Zhou, Bo, Baojiang Zheng, Weilin Wu. "Environmental Stress-Induced Anthocyanin Accumulation in Plants" Encyclopedia, https://encyclopedia.pub/entry/55069 (accessed February 08, 2026).
Zhou, B., Zheng, B., & Wu, W. (2024, February 15). Environmental Stress-Induced Anthocyanin Accumulation in Plants. In Encyclopedia. https://encyclopedia.pub/entry/55069
Zhou, Bo, et al. "Environmental Stress-Induced Anthocyanin Accumulation in Plants." Encyclopedia. Web. 15 February, 2024.
Copy Citation
Plants have evolved complicated defense and adaptive systems to grow in various abiotic stress environments such as drought, cold, and salinity. Anthocyanins belong to the secondary metabolites of flavonoids with strong antioxidant activity in response to various abiotic stress and enhance stress tolerance. Anthocyanin accumulation often accompanies the resistance to abiotic stress in plants to scavenge reactive oxygen species (ROS).
anthocyanin biosynthesis
ncRNA
environmental regulation
abiotic stress
1. Introduction
With population growth and environmental deterioration, plants face more and more abiotic stresses. As sessile organisms, they are exposed to various abiotic stresses during their growth and development process, and they have also evolved sophisticated tolerance, resistance, or avoidance mechanisms to overcome stress such as low and high temperature, drought, and salinity [1][2]. Under abiotic stress, the stress stimuli are perceived by receptors or signal factors and transduced to downstream transcription factors through messengers like calcium (Ca2+), nitric oxide (NO), sugars, abscisic acid (ABA), brassinosteroids (BRs), ethylene, jasmonates (JA), salicylic acid (SA), and auxins in plants [3]. Under abiotic stresses, plants usually generate and accumulate secondary metabolites including anthocyanins, flavones, flavonols, lignin, alkaloids, and terpenoids to protect cells from damage [4][5].
Anthocyanins are a class of water-soluble flavonoids present in fruits, flowers, and vegetative organs of plants which have antioxidant activities in protecting plants under abiotic stress. Anthocyanidins are synthesized on the cytoplasmic surface of the endoplasmic reticulum and further undergo various modifications such as methylation, glycosylation, hydroxylation, and acylation in the endoplasmic reticulum. Afterward, anthocyanins (the forms of anthocyanidin glycosides and acylated anthocyanins) enter the vacuoles to store and accumulate with the assistance of transporters and transport vesicles [6]. Anthocyanins not only determine the blue, red, and purple pigments of plants for attracting pollinators but also play roles in various biotic and abiotic stresses. In addition to physiological roles for plants, anthocyanins also have potential benefits for human health, such as decreasing the risk of heart disease, diabetes, cardiovascular disease, and metabolic diseases [7][8][9][10]. The anthocyanin biosynthetic pathway has been elucidated and most of the regulatory genes involved in anthocyanin biosynthesis have been identified [11][12][13]. The enzymes involved in anthocyanin biosynthesis include CHS (chalcone synthase), CHI (chalcone isomerase), F3H (flavanone 3-hydroxylase), F3′H (flavonoid 3′-hydroxylase), F3′5′H (flavonoid 3′,5′-hydroxylase) which correspond to early biosynthetic genes (EBGs), and DFR (dihydroflavonol 4-reductase), ANS (anthocyanin synthase), OMT (O-methyltransferase) and UFGT (UDP flavonoid glucosyltransferase) which correspond to late biosynthetic genes (LBGs) [14]. Moreover, the anthocyanin biosynthesis is regulated by the MYB–bHLH–WD40 (MBW) protein complex, which is composed of MYB, bHLH transcription factors, and a WD40 protein [15][16]. In addition to the MBW complex, PybHLH3-PyMYB114-PyERF3 [17] transcription complex in pear and WRKY [18][19], NAC [20], MADS [21], HY5 [22], BBX [23], bZIP [24], SPL [25] regulatory factors in various fruit crops are also involved in the regulation of anthocyanin biosynthesis (Figure 1). Recently, a great number of studies have revealed that anthocyanins increasingly accumulate when plants are under environmental stress. In Arabidopsis, the low nitrogen (N)-induced anthocyanin accumulation plays a substantial role in plant tolerance to low N stress [26]. In addition, the flavonoid biosynthesis and accumulation in Arabidopsis improves salt resistance under salt stress [27]. In Cymbidium hybrid flowers, anthocyanin pigmentation has been demonstrated to be organ-specific and temperature-dependent synthesized [28]. Moreover, anthocyanin accumulation is related to salt stress response in MdZAT5-overexpressing apple Calli and Arabidopsis [29]. Furthermore, the anthocyanin accumulation in AN1-overexpressing tobacco plants has a higher drought tolerance compared to the wild-type plants [30]. Also, the overexpression of UGTs enhanced plant tolerance to low temperatures, drought, and salt stresses by modulating the anthocyanin accumulation [31]. Under abiotic stresses, a reduction in electron transport in the Calvin cycle and a higher electron leakage during photosynthesis in the Mehler reaction of cells lead to the plants producing extensive reactive oxygen species (ROS) including O2−, H2O2, OH-, and 1O2 [32], which cause oxidative damage to plants and can be used as signaling molecules to activate stress-tolerance mechanisms. Anthocyanin can directly scavenge active oxygen species, such as singlet oxygen, superoxide, hydrogen peroxide, hydroxyl, and peroxyl radicals [33]. Consequently, the anthocyanins accumulate in plants after ROS signals induce the transcription of anthocyanin biosynthesis pathway genes to scavenge excess ROS and avoid oxidative damage [34][35]. Therefore, ROS as signal factors in the response to plant abiotic stresses activate the transcription of anthocyanin biosynthetic genes to produce anthocyanins for stress tolerance.
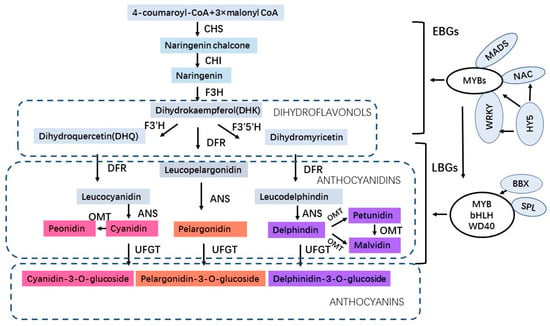
Figure 1. The regulatory pathway of anthocyanin biosynthesis.
2. Environmental Stress-Induced Anthocyanin Accumulation in Plants
2.1. The Pathway of Transcription Factors and ncRNAs Involved in Low Temperature Stress Response
Low temperature is one of the major abiotic stresses that greatly reduces crop yield. The ability of plants to tolerate adverse environments is known to affect plant survival and geographical distribution, for example, plants growing in temperate zones are less sensitive to cold than those from tropical/subtropical regions, such as rice, maize, cotton, and tomatoes, which cannot adapt to cold environments [36]. Cold stress can be classified into chilling stress (0–15 °C) and freezing stress (<0 °C), which can cause injuries to the plant. However, some plants such as winter wheat (Triticum aestivum), rye (Secale cereale L.), barley (Hordeum vulgare), and oat (Avena sativa) have evolved sophisticated cold acclimation mechanisms to encounter low temperatures that improve plant freezing tolerance [37].
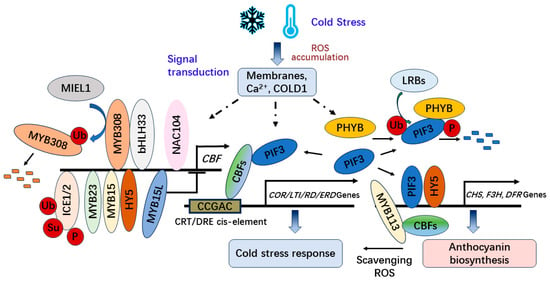
Currently, the cold signal has been reported to be perceived by cellular membranes, calcium (Ca2+) channels, and COLD1 (CHILLING TOLERANCE DIVERGENCE 1), which encodes a G-protein signaling regulator in Oryza sativa [38]. Afterwards, the cold signal is transduced to the C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 (CBF/DREB1)-dependent regulatory pathway for plant responses to cold stress. Additionally, the circadian clock, photoperiod, and light signaling are involved in regulating the network [38]. CBF proteins recognize the conserved CCGAC sequence of CRT/DRE cis-element in the promoters of a subset of cold-regulated (COR) genes [39]. The cold-regulated genes including the COR, low-temperature induced (LTI), responsive to desiccation (RD), and early dehydration-inducible (ERD) genes can be activated to increase freezing tolerance through the regulation of the CBF/DREB1 transcription factors in the cold acclimation of plants [37]. During the cold signal transduction, ICE1 (INDUCER OF CBF EXPRESSION 1) and its homolog ICE2 positively regulate CBF expression, and the modifications of post-translation in ICE1 such as ubiquitination, sumoylation, and phosphorylation attenuate the activity and stability of ICE1 to affect the expression abundance of CBF [37][38] (Figure 2).

Figure 2. The CBF-dependent regulation of anthocyanin biosynthesis and cold tolerance under cold stress.
Furthermore, CBF can interact with PHYTOCHROME-INTERACTING FACTOR 3 (PIF3) under cold stress attenuating the mutually assured destruction of PIF3-phyB to stabilize PHYTOCHROME B (phyB) and positively regulate freezing tolerance by modulating the expression of stress-responsive genes in Arabidopsis [40]. In addition, PIF3 can be ubiquitinated by LRBs (LIGHT-RESPONSE BRIC-A-BRACK/TRAMTRACK/BROAD) for degradation after interacting with PhyB to be phosphorylated [41][42][43][44].
Moreover, CBFs in eggplants interact with SmMYB113, a key regulator of anthocyanin biosynthesis, to upregulate the expression of CHS and DFR with a SmMYB113-dependent pathway to improve the contents of anthocyanin. Overexpression of SmCBF2 and SmCBF3 in Arabidopsis also enhances the anthocyanin accumulation under cold conditions [45].
Moreover, the NAC (NAM, ATAF1/2, and CUC2) transcription factor MdNAC104 in apples also promotes the expression of MdCBF1 and MdCBF3, the anthocyanin synthesis-related genes such as MdCHS-b, MdCHI-a, MdF3H-a and MdANS-b and the antioxidant enzyme-encoding genes MdFSD2 and MdPRXR1.1 under cold stress [46].
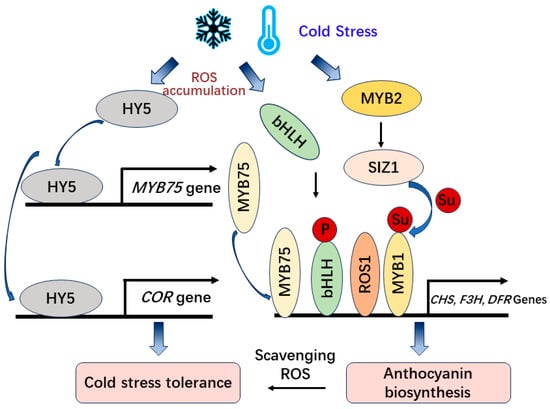
Apart from the CBF-dependent regulatory pathway, a number of genes have been identified to regulate plant cold responses independently of the CBF pathway (Figure 3). In Arabidopsis, cold stress induces the expression of HY5 (ELONGATED HYPOCOTYL 5) and positively regulates the expression of COR genes and cold acclimation via a CBF-independent pathway [47]. Moreover, HY5 has been determined to regulate anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis [48] and regulating the expression of anthocyanin biosynthetic genes (CHS, CHI, and F3H) in tomato under cold stress [49].

Figure 3. The CBF-independent regulation of anthocyanin biosynthesis and cold tolerance under cold stress.
Low temperature is a serious abiotic stressor that induces ROS (reactive oxygen species), which participate in stress signaling as signaling molecules through the protein kinases pathway to activate anthocyanin regulatory transcription factors such as MYB, bHLH, and WD40 [50][51]. The accumulation of anthocyanins in plant leaves at low temperatures can decrease the level of oxidative damage and increase the photosynthetic rate [52]. In Mikania micrantha, the accumulation of anthocyanins in leaves and stems could effectively eliminate ROS under low-temperature stress and improve its adaptability to low-temperature environments during winter [53]. The anthocyanins scavenging ROS and maintaining osmotic balance to increase abiotic stress tolerance has also been proved using anthocyanin-containing leaves compared with anthocyanin-deficient leaves in sweet basil (Ocimum basilicum) and Arabidopsis [51][54]. Also, cold-induced transcription and phosphorylation of bHLH activate the expression of MdDFR, MdUFGT, and MdMYB1 to regulate anthocyanin accumulation and fruit coloration in apples [55]. Additionally, low temperature induces MdMYB2 and further activates the expression of MdSIZ1 (SAP AND MIZ1 DOMAIN-CONTAINING LIGASE1), which is SUMO E3 ligase that mediates the sumoylation of MdMYB1 and promotes anthocyanin accumulation [56][57] (Figure 3).
Non-coding RNAs also play crucial roles in the plant’s response to cold stress. An overexpression of MIR156 in tobacco leaves for transient expression assay showed higher anthocyanin levels and an enhanced cold stress tolerance than that in the control. Additionally, the lower extent of H2O2 accumulation and the higher expression of early responsive dehydration (ERD) gene family, ERD10B, ERD10C, and ERD10D, which are downstream genes in the ICE-CBF-COR pathway, were detected under cold stress [58]. SPL9, one of the miR156 targets, has been reported to inhibit the expression of anthocyanin synthesis genes by competitively interacting with PAP1 (production of the anthocyanin pigments1), the component of the MYB–BHLH–WD40 complex, resulting in the interference with the regulation of anthocyanin accumulation [59]. The negative regulation of SPL9 targeted by miR156 caused the accumulation of anthocyanins in the MIR156-overexpressing tobacco leaves preventing the overproduction and accumulation of ROS in tobacco leaves. The miR828/858-MYBs factors are also related to regulating needle discoloration of C. fortunei in cold winters [60].
Furthermore, miR397 overexpression in Arabidopsis improves tolerance to cold stress by inducing high levels of CBF and COR by targeting LACs (Laccase) and CKB3 (Casein Kinase II Subunit Beta 3) in the cold signaling pathway [61]. Whereas the lncRNA, FRILAIR (FRUIT RIPENING-RELATED LONG INTERGENIC RNA) acts as a noncanonical target mimic of miR397 to modulate the expression of LAC11a (encoding a putative laccase-11-like protein), which promotes expressions of anthocyanin biosynthetic genes in the strawberry fruit ripening process [62].
In addition, MdLNC499 can target MdERF109 to regulate light-induced anthocyanin accumulation in apple fruit [63] and the ERF109 in trifoliate orange also contributes to cold tolerance by regulating Prx1 involved in the antioxidative process [64]. Another lncRNA, MdLNC610, upregulates the expression of MdACO1 and participates in high-light-induced ethylene and anthocyanin biosynthesis [65].
2.2. High-Temperature Stress Response Factors
Floral pigmentation has been reported to respond differently to changes in temperature, and plants with exposed anthers increased in pigmentation with temperature increasing whereas plants with concealed anthers declined in pigmentation [66]. Distinct from the response of low temperature, plants generate flavones to eliminate excess ROS at short-term high-temperature conditions in colorless plant organs. However, R2R3-MYB transcription factor CmMYB012 was found to respond to long-time high-temperature stress and inhibit flavonoid biosynthesis by directly regulating flavone synthase in the chrysanthemum [67].
High temperature also induces the degradation of the HY5 protein in a COP1 activity-dependent manner and the degradation of HY5 derepresses the expression of MYBL2, which partially mediates the high-temperature repression of anthocyanin biosynthesis in Arabidopsis [68]. Furthermore, HY5 activates miR858a, which targets the anthocyanin repressor MYBL2 and increases anthocyanin biosynthesis in Arabidopsis [69]. In apples, MdHY5 promotes anthocyanin accumulation by regulating the expression of the MdMYB10 gene and downstream anthocyanin biosynthesis genes [22] and MdHY5 also inhibits the transcription of mdm-miR858 which targets the transcription factor genes MdMYB9 and MdMYBPA1 to upregulate anthocyanin accumulation [70].
Additionally, anthocyanin concentration depends on the counterbalance between its synthesis and degradation in plum fruit at high temperatures. The high temperature increased the concentration of hydrogen peroxide and the activity of class III peroxidase in the fruit. The hydrogen peroxide degraded anthocyanin, while class III peroxidase catalyzed hydrogen peroxide [71].
Consequently, plants exposed to heat stress benefit from a decrease in anthocyanin because of the activation of negative factors, the degradation of positive regulators in anthocyanin biosynthesis, the high concentration of hydrogen peroxide, and the low level of ABA to degrade and reduce anthocyanin accumulation. The decreased anthocyanin accumulation under heat stress is caused by the transcriptional reduction of both early and late anthocyanin biosynthetic genes which are regulated by positive and negative regulators [69] and the acceleration of anthocyanin degradation [72].
Plants have also evolved salt tolerance mechanisms through regulatory genes under salt stress. In torenia “Kauai Rose”, overexpression of anthocyanin regulatory transcription factors (RsMYB1 or B-Peru + mPAP1) can alleviate salt stress-induced growth inhibition by reducing the salt stress-induced ROS and MDA contents [73]. In Arabidopsis, MYB3 functions as a transcriptional repressor for the regulation of lignin and anthocyanin biosynthesis under high salt conditions [74]. The myb3 mutant plants exhibited high accumulation of lignin and anthocyanin and longer root growth in high NaCl conditions than wild-type plants and anthocyanin biosynthetic genes, such as phenylalanine ammonia-lyase 1 (PAL1), cinnamate 4-hydroxylase (C4H), catechol-O-methyltransferase (COMT), 4-coumaric acid-CoA ligase (4CL), dihydroflavonol reductase (DFR), and leucoanthocyanidin dioxygenase (LDOX) were also upregulated [74].
Many miRNA target modules such as miR156-SPL, miR160-ARF, miR171-SCL (SCARECROW-LIKE), miR172-AP2, miR319-TCP, miR390-TAS3-ARF, miR393-TIR1/AFB, miR394-LCR, miR395-APS/SULTR2;1, miR396-GRF, miR397-LAC, miR398-CSD, miR399-PHO2, miR408-LAC/PLANTACYANIN, miR414-FSD1, miR528-AAO/LAC, miR535-SPL, and miRNAs such as miR402, miR417, miR1861h, miRNVL5 participate in plant salt stress responses by regulating hormonal signaling pathways and antioxidant system [75].
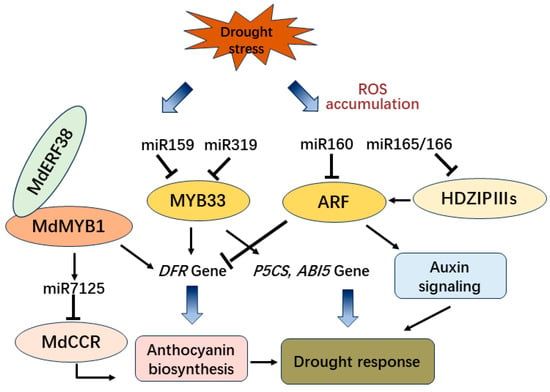
2.4. The Transcription Factors and ncRNAs Involved in Drought Stress-Induced Anthocyanin Biosynthesis
Drought stress is one of the most common stresses in plants. When the water supply to the roots is limited or the transpiration rate is too high, the limited water supply leads to an imbalance between light capture and photosynthesis, allowing oxidative stress to occur. Drought stress induces reactive oxygen species (ROS), such as hydroxyl radicals (·OH), superoxide radicals (O2•−), and hydrogen peroxide (H2O2), and anthocyanins exhibit strong radical-scavenging activity avoiding excess ROS accumulation to cause cell death [76].
Under drought conditions, overexpression of BoNAC019 from Brassica oleracea decreased the content of antioxidant enzymes and anthocyanin which conversely results in the accumulation of more reactive oxygen species (ROS). Moreover, the expression of antioxidant enzymatic genes, anthocyanin biosynthetic genes, and ABA signaling genes were also downregulated [77]. Moreover, in Arabidopsis, the loss-of-function of miR159 enhanced drought tolerance and hypersensitivity of seed germination to ABA, while MYB33, the target of miR159, acts upstream of ABI5 in the ABA signaling pathway to regulate drought response and seed germination in plants [78]. Furthermore, the overexpression of MYB65 and MYB101 in Arabidopsis, with a mutated recognition site in miR159, causes hypersensitivity to ABA and a relatively high tolerance to drought conditions [79]. However, in cotton, GhMYB33 targeted by miR319c controls the transcription of GhDFR1 to promote the accumulation of anthocyanin [80] (Figure 4).

Figure 4. The regulation pathway of miRNAs and transcription factors involved in drought tolerance and anthocyanin biosynthesis.
The expression of Stu-miR159 decreased in response to drought treatment, while the expression of StGAMyb-like target genes increased with drought stress in potatoes [81]. Similarly, in tomatoes, the expression of sly-miR159 decreases in response to drought stress, and its target SlMYB33 correlates with the induction of SlP5CS gene expression and accumulation of the osmoprotective compounds proline and putrescine [82]. Moreover, in mulberry, mul-miR159a plays a negative regulatory role in the biosynthesis of anthocyanins by targeting the Mul-MYB33 gene [83]. Therefore, the decreasing expression of miR159 under drought stress might be related to the accumulation of anthocyanin in plants.
Additionally, in Arabidopsis, STTM165/166 promotes the expression of HD-ZIP IIIs, which also activates the expression of ARFs (AUXIN RESPONSE FACTORS) targeted by miR160 and shows tolerance to drought stress [84]. Moreover, miR160h-ARF18 was identified as potentially controlling the accumulation of anthocyanins in poplar [5].
2.5. Long ncRNAs Related to Anthocyanin Synthesis
LncRNA can act as a regulator to affect the expression of genes involved in the anthocyanin biosynthetic pathway. For instance, MdLNC499, a long non-coding RNA regulated by MdWRKY1, induces the expression of MdERF109, which is involved in the light-induced anthocyanin synthesis pathway in apple fruit [63]. Moreover, MdLNC610, a positive regulator of MdACO1 in ethylene biosynthesis, is also involved in the regulation of anthocyanin production induced by the high-light intensity in apple (Malus domestica) [65].
3. Conclusions
Abiotic stress has extremely inhibited plant growth and crop productivity with the ongoing deterioration of the global climate and environment. However, plants have also evolved the defense mechanism to adapt to various stresses. Stresses induce the production of reactive oxygen species (ROS) and over-accumulation of ROS causes oxidative damage to plants. Simultaneously, anthocyanins are major bioactive compounds induced by abiotic stress as potent antioxidants to scavenge ROS [35]. Under cold stress, CBFs-dependent and -independent regulation of anthocyanin biosynthesis and COR/LTI/RD/ERD genes take roles in plant cold tolerance. However, the high temperature usually decreases anthocyanin accumulation and enhances tolerance to heat stress through regulating the expression of HSFs and HSPs. Moreover, salt stress induces MYB transcription factors and miRNAs involved in the regulation of anthocyanin and antioxidant enzymes for ROS clearance to increase stress tolerance.
References
- Kumar, A.A.; Mishra, P.; Kumari, K.; Panigrahi, K.C. Environmental stress influencing plant development and flowering. Front. Biosci. (Schol. Ed.) 2012, 4, 1315–1324.
- Chen, Z.H.; Soltis, D.E. Evolution of environmental stress responses in plants. Plant Cell Environ. 2020, 43, 2827–2831.
- Kaur, N.; Gupta, A.K. Signal transduction pathways under abiotic stresses in plants. Curr. Sci. India 2005, 88, 1771–1780.
- Dong, N.Q.; Lin, H.X. Contribution of phenylpropanoid metabolism to plant development and plant-environment interactions. J. Integr. Plant Biol. 2021, 63, 180–209.
- Wang, Y.; Liu, W.; Wang, X.; Yang, R.; Wu, Z.; Wang, H.; Wang, L.; Hu, Z.; Guo, S.; Zhang, H.; et al. MiR156 regulates anthocyanin biosynthesis through SPL targets and other microRNAs in poplar. Hortic. Res. 2020, 7, 118.
- Han, L.; Zhou, L.; Zou, H.; Yuan, M.; Wang, Y. PsGSTF3, an Anthocyanin-Related Glutathione S-Transferase Gene, Is Essential for Petal Coloration in Tree Peony. Int. J. Mol. Sci. 2022, 23, 1423.
- Wallace, T.C. Anthocyanins in cardiovascular disease. Adv. Nutr. 2011, 2, 1–7.
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779.
- Lila, M.A. Anthocyanins and Human Health: An In Vitro Investigative Approach. J. Biomed. Biotechnol. 2004, 2004, 306–313.
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A Comprehensive Review of Their Chemical Properties and Health Effects on Cardiovascular and Neurodegenerative Diseases. Molecules 2020, 25, 3809.
- Sunil, L.; Shetty, N.P. Biosynthesis and regulation of anthocyanin pathway genes. Appl. Microbiol. Biotechnol. 2022, 106, 1783–1798.
- Ma, Y.; Ma, X.; Gao, X.; Wu, W.; Zhou, B. Light Induced Regulation Pathway of Anthocyanin Biosynthesis in Plants. Int. J. Mol. Sci. 2021, 22, 11116.
- Tang, B.; Li, L.; Hu, Z.; Chen, Y.; Tan, T.; Jia, Y.; Xie, Q.; Chen, G. Anthocyanin Accumulation and Transcriptional Regulation of Anthocyanin Biosynthesis in Purple Pepper. J. Agric. Food Chem. 2020, 68, 12152–12163.
- Liu, Y.; Tikunov, Y.; Schouten, R.E.; Marcelis, L.F.M.; Visser, R.G.F.; Bovy, A. Anthocyanin Biosynthesis and Degradation Mechanisms in Solanaceous Vegetables: A Review. Front. Chem. 2018, 6, 52.
- Xu, W.; Dubos, C.; Lepiniec, L. Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci. 2015, 20, 176–185.
- Li, S. Transcriptional control of flavonoid biosynthesis: Fine-tuning of the MYB-bHLH-WD40 (MBW) complex. Plant Signal. Behav. 2014, 9, e27522.
- Yao, G.; Ming, M.; Allan, A.C.; Gu, C.; Li, L.; Wu, X.; Wang, R.; Chang, Y.; Qi, K.; Zhang, S.; et al. Map-based cloning of the pear gene MYB114 identifies an interaction with other transcription factors to coordinately regulate fruit anthocyanin biosynthesis. Plant J. 2017, 92, 437–451.
- Li, C.; Wu, J.; Hu, K.D.; Wei, S.W.; Sun, H.Y.; Hu, L.Y.; Han, Z.; Yao, G.F.; Zhang, H. PyWRKY26 and PybHLH3 cotargeted the PyMYB114 promoter to regulate anthocyanin biosynthesis and transport in red-skinned pears. Hortic. Res. 2020, 7, 37.
- An, J.P.; Zhang, X.W.; You, C.X.; Bi, S.Q.; Wang, X.F.; Hao, Y.J. MdWRKY40 promotes wounding-induced anthocyanin biosynthesis in association with MdMYB1 and undergoes MdBT2-mediated degradation. New Phytol. 2019, 224, 380–395.
- Zhou, H.; Lin-Wang, K.; Wang, H.; Gu, C.; Dare, A.P.; Espley, R.V.; He, H.; Allan, A.C.; Han, Y. Molecular genetics of blood-fleshed peach reveals activation of anthocyanin biosynthesis by NAC transcription factors. Plant J. 2015, 82, 105–121.
- Jaakola, L.; Poole, M.; Jones, M.O.; Kamarainen-Karppinen, T.; Koskimaki, J.J.; Hohtola, A.; Haggman, H.; Fraser, P.D.; Manning, K.; King, G.J.; et al. A SQUAMOSA MADS box gene involved in the regulation of anthocyanin accumulation in bilberry fruits. Plant Physiol. 2010, 153, 1619–1629.
- An, J.P.; Qu, F.J.; Yao, J.F.; Wang, X.N.; You, C.X.; Wang, X.F.; Hao, Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017, 4, 17023.
- Bai, S.; Tao, R.; Tang, Y.; Yin, L.; Ma, Y.; Ni, J.; Yan, X.; Yang, Q.; Wu, Z.; Zeng, Y.; et al. BBX16, a B-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 2019, 17, 1985–1997.
- Liu, H.; Su, J.; Zhu, Y.; Yao, G.; Allan, A.C.; Ampomah-Dwamena, C.; Shu, Q.; Lin-Wang, K.; Zhang, S.; Wu, J. The involvement of PybZIPa in light-induced anthocyanin accumulation via the activation of PyUFGT through binding to tandem G-boxes in its promoter. Hortic. Res. 2019, 6, 134.
- Liu, H.; Shu, Q.; Lin-Wang, K.; Allan, A.C.; Espley, R.V.; Su, J.; Pei, M.; Wu, J. The PyPIF5-PymiR156a-PySPL9-PyMYB114/MYB10 module regulates light-induced anthocyanin biosynthesis in red pear. Mol. Hortic. 2021, 1, 14.
- Liang, J.; He, J.X. Protective role of anthocyanins in plants under low nitrogen stress. Biochem. Biophys. Res. Commun. 2018, 498, 946–953.
- Li, B.Z.; Fan, R.N.; Guo, S.Y.; Wang, P.T.; Zhu, X.H.; Fan, Y.T.; Chen, Y.X.; He, K.Y.; Kumar, A.; Shi, J.P.; et al. The Arabidopsis MYB transcription factor, MYB111 modulates salt responses by regulating flavonoid biosynthesis. Environ. Exp. Bot. 2019, 166, 103807.
- Nakatsuka, T.; Suzuki, T.; Harada, K.; Kobayashi, Y.; Dohra, H.; Ohno, H. Floral organ- and temperature-dependent regulation of anthocyanin biosynthesis in Cymbidium hybrid flowers. Plant Sci. 2019, 287, 110173.
- Wang, D.R.; Yang, K.; Wang, X.; Lin, X.L.; Rui, L.; Liu, H.F.; Liu, D.D.; You, C.X. Overexpression of MdZAT5, an C2H2-Type Zinc Finger Protein, Regulates Anthocyanin Accumulation and Salt Stress Response in Apple Calli and Arabidopsis. Int. J. Mol. Sci. 2022, 23, 1897.
- Cirillo, V.; D’Amelia, V.; Esposito, M.; Amitrano, C.; Carillo, P.; Carputo, D.; Maggio, A. Anthocyanins are Key Regulators of Drought Stress Tolerance in Tobacco. Biology 2021, 10, 139.
- Li, P.; Li, Y.J.; Zhang, F.J.; Zhang, G.Z.; Jiang, X.Y.; Yu, H.M.; Hou, B.K. The Arabidopsis UDP-glycosyltransferases UGT79B2 and UGT79B3, contribute to cold, salt and drought stress tolerance via modulating anthocyanin accumulation. Plant J. 2017, 89, 85–103.
- Mittler, R. ROS Are Good. Trends Plant Sci. 2017, 22, 11–19.
- Kaur, S.; Tiwari, V.; Kumari, A.; Chaudhary, E.; Sharma, A.; Ali, U.; Garg, M. Protective and defensive role of anthocyanins under plant abiotic and biotic stresses: An emerging application in sustainable agriculture. J. Biotechnol. 2023, 361, 12–29.
- Qin, L.; Sun, L.; Wei, L.; Yuan, J.; Kong, F.; Zhang, Y.; Miao, X.; Xia, G.; Liu, S. Maize SRO1e represses anthocyanin synthesis through regulating the MBW complex in response to abiotic stress. Plant J. 2021, 105, 1010–1025.
- Naing, A.H.; Kim, C.K. Abiotic stress-induced anthocyanins in plants: Their role in tolerance to abiotic stresses. Physiol. Plant. 2021, 172, 1711–1723.
- Thomashow, M.F. PLANT COLD ACCLIMATION: Freezing Tolerance Genes and Regulatory Mechanisms. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 1999, 50, 571–599.
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637.
- Ding, Y.; Shi, Y.; Yang, S. Molecular Regulation of Plant Responses to Environmental Temperatures. Mol. Plant 2020, 13, 544–564.
- Du, Z.; Li, J. Expression, purification and molecular characterization of a novel transcription factor KcCBF3 from Kandelia candel. Protein Expr. Purif. 2019, 153, 26–34.
- Jiang, B.; Shi, Y.; Peng, Y.; Jia, Y.; Yan, Y.; Dong, X.; Li, H.; Dong, J.; Li, J.; Gong, Z.; et al. Cold-Induced CBF-PIF3 Interaction Enhances Freezing Tolerance by Stabilizing the phyB Thermosensor in Arabidopsis. Mol. Plant 2020, 13, 894–906.
- Ni, W.; Xu, S.L.; Gonzalez-Grandio, E.; Chalkley, R.J.; Huhmer, A.F.R.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 2017, 8, 15236.
- Ni, W.; Xu, S.L.; Tepperman, J.M.; Stanley, D.J.; Maltby, D.A.; Gross, J.D.; Burlingame, A.L.; Wang, Z.Y.; Quail, P.H. A mutually assured destruction mechanism attenuates light signaling in Arabidopsis. Science 2014, 344, 1160–1164.
- Favero, D.S. Mechanisms regulating PIF transcription factor activity at the protein level. Physiol. Plant. 2020, 169, 325–335.
- Kim, C.; Kwon, Y.; Jeong, J.; Kang, M.; Lee, G.S.; Moon, J.H.; Lee, H.J.; Park, Y.I.; Choi, G. Phytochrome B photobodies are comprised of phytochrome B and its primary and secondary interacting proteins. Nat. Commun. 2023, 14, 1708.
- Zhou, L.; He, Y.; Li, J.; Liu, Y.; Chen, H. CBFs Function in Anthocyanin Biosynthesis by Interacting with MYB113 in Eggplant (Solanum melongena L.). Plant Cell Physiol. 2020, 61, 416–426.
- Mei, C.; Yang, J.; Mei, Q.; Jia, D.; Yan, P.; Feng, B.; Mamat, A.; Gong, X.; Guan, Q.; Mao, K.; et al. MdNAC104 positively regulates apple cold tolerance via CBF-dependent and CBF-independent pathways. Plant Biotechnol. J. 2023, 21, 2057–2073.
- Catala, R.; Medina, J.; Salinas, J. Integration of low temperature and light signaling during cold acclimation response in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 16475–16480.
- Shin, D.H.; Choi, M.; Kim, K.; Bang, G.; Cho, M.; Choi, S.B.; Choi, G.; Park, Y.I. HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 2013, 587, 1543–1547.
- Han, N.; Fan, S.; Zhang, T.; Sun, H.; Zhu, Y.; Gong, H.; Guo, J. SlHY5 is a necessary regulator of the cold acclimation response in tomato. Plant Growth Regul. 2020, 91, 1–12.
- Altangerel, N.; Ariunbold, G.O.; Gorman, C.; Alkahtani, M.H.; Borrego, E.J.; Bohlmeyer, D.; Hemmer, P.; Kolomiets, M.V.; Yuan, J.S.; Scully, M.O. In vivo diagnostics of early abiotic plant stress response via Raman spectroscopy. Proc. Natl. Acad. Sci. USA 2017, 114, 3393–3396.
- Xu, Z.; Mahmood, K.; Rothstein, S.J. ROS Induces Anthocyanin Production Via Late Biosynthetic Genes and Anthocyanin Deficiency Confers the Hypersensitivity to ROS-Generating Stresses in Arabidopsis. Plant Cell Physiol. 2017, 58, 1364–1377.
- Zhu, H.; Zhang, T.J.; Zheng, J.; Huang, X.D.; Yu, Z.C.; Peng, C.L.; Chow, W.S. Anthocyanins function as a light attenuator to compensate for insufficient photoprotection mediated by nonphotochemical quenching in young leaves of Acmena acuminatissima in winter. Photosynthetica 2018, 56, 445–454.
- Zhang, Q.; Zhai, J.; Shao, L.; Lin, W.; Peng, C. Accumulation of Anthocyanins: An Adaptation Strategy of Mikania micrantha to Low Temperature in Winter. Front. Plant Sci. 2019, 10, 1049.
- Landi, M.; Guidi, L.; Pardossi, A.; Tattini, M.; Gould, K.S. Photoprotection by foliar anthocyanins mitigates effects of boron toxicity in sweet basil (Ocimum basilicum). Planta 2014, 240, 941–953.
- Xie, X.B.; Li, S.; Zhang, R.F.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897.
- Jiang, H.; Zhou, L.J.; Gao, H.N.; Wang, X.F.; Li, Z.W.; Li, Y.Y. The transcription factor MdMYB2 influences cold tolerance and anthocyanin accumulation by activating SUMO E3 ligase MdSIZ1 in apple. Plant Physiol. 2022, 189, 2044–2060.
- Zhou, L.J.; Li, Y.Y.; Zhang, R.F.; Zhang, C.L.; Xie, X.B.; Zhao, C.; Hao, Y.J. The small ubiquitin-like modifier E3 ligase MdSIZ1 promotes anthocyanin accumulation by sumoylating MdMYB1 under low-temperature conditions in apple. Plant Cell Environ. 2017, 40, 2068–2080.
- Yang, Y.; Zhang, X.; Su, Y.; Zou, J.; Wang, Z.; Xu, L.; Que, Y. miRNA alteration is an important mechanism in sugarcane response to low-temperature environment. BMC Genom. 2017, 18, 833.
- Gou, J.Y.; Felippes, F.F.; Liu, C.J.; Weigel, D.; Wang, J.W. Negative regulation of anthocyanin biosynthesis in Arabidopsis by a miR156-targeted SPL transcription factor. Plant Cell 2011, 23, 1512–1522.
- Zhang, Y.; Yang, J.; Zhu, L.; Xue, J.; Hu, H.; Cui, J.; Xu, J. Identification of microRNAs and their target genes related to needle discoloration of evergreen tree Chinese cedar (Cryptomeria fortunei) in cold winters. Planta 2021, 254, 31.
- Dong, C.-H.; Pei, H. Over-expression of miR397 improves plant tolerance to cold stress in Arabidopsis thaliana. J. Plant Biol. 2014, 57, 209–217.
- Tang, Y.; Qu, Z.; Lei, J.; He, R.; Adelson, D.L.; Zhu, Y.; Yang, Z.; Wang, D. The long noncoding RNA FRILAIR regulates strawberry fruit ripening by functioning as a noncanonical target mimic. PLoS Genet. 2021, 17, e1009461.
- Ma, H.; Yang, T.; Li, Y.; Zhang, J.; Wu, T.; Song, T.; Yao, Y.; Tian, J. The long noncoding RNA MdLNC499 bridges MdWRKY1 and MdERF109 function to regulate early-stage light-induced anthocyanin accumulation in apple fruit. Plant Cell 2021, 33, 3309–3330.
- Wang, M.; Dai, W.; Du, J.; Ming, R.; Dahro, B.; Liu, J.H. ERF109 of trifoliate orange (Poncirus trifoliata (L.) Raf.) contributes to cold tolerance by directly regulating expression of Prx1 involved in antioxidative process. Plant Biotechnol. J. 2019, 17, 1316–1332.
- Yu, J.; Qiu, K.; Sun, W.; Yang, T.; Wu, T.; Song, T.; Zhang, J.; Yao, Y.; Tian, J. A long non-coding RNA functions in high-light-induced anthocyanin accumulation in apple by activating ethylene synthesis. Plant Physiol. 2022, 189, 66–83.
- Koski, M.H.; MacQueen, D.; Ashman, T.-L. Floral Pigmentation Has Responded Rapidly to Global Change in Ozone and Temperature. Curr. Biol. 2020, 30, 4425–4431.E3.
- Zhou, L.J.; Geng, Z.; Wang, Y.; Wang, Y.; Liu, S.; Chen, C.; Song, A.; Jiang, J.; Chen, S.; Chen, F. A novel transcription factor CmMYB012 inhibits flavone and anthocyanin biosynthesis in response to high temperatures in chrysanthemum. Hortic. Res. 2021, 8, 248.
- Kim, S.; Hwang, G.; Lee, S.; Zhu, J.Y.; Paik, I.; Nguyen, T.T.; Kim, J.; Oh, E. High Ambient Temperature Represses Anthocyanin Biosynthesis through Degradation of HY5. Front. Plant Sci. 2017, 8, 1787.
- Wang, Y.; Wang, Y.; Song, Z.; Zhang, H. Repression of MYBL2 by Both microRNA858a and HY5 Leads to the Activation of Anthocyanin Biosynthetic Pathway in Arabidopsis. Mol. Plant 2016, 9, 1395–1405.
- Li, Z.; Liu, W.; Chen, Q.; Zhang, S.; Mei, Z.; Yu, L.; Wang, C.; Mao, Z.; Chen, Z.; Chen, X.; et al. Mdm-miR858 targets MdMYB9 and MdMYBPA1 to participate anthocyanin biosynthesis in red-fleshed apple. Plant J. 2023, 113, 1295–1309.
- Niu, J.; Zhang, G.; Zhang, W.; Goltsev, V.; Sun, S.; Wang, J.; Li, P.; Ma, F. Anthocyanin concentration depends on the counterbalance between its synthesis and degradation in plum fruit at high temperature. Sci. Rep. 2017, 7, 7684.
- Tan, Y.; Wen, B.; Xu, L.; Zong, X.; Sun, Y.; Wei, G.; Wei, H. High temperature inhibited the accumulation of anthocyanin by promoting ABA catabolism in sweet cherry fruits. Front. Plant Sci. 2023, 14, 1079292.
- Zhao, R.; Naing, A.H.; Xu, J.; Kim, C.K. Overexpression of anthocyanin regulatory transcription factors can alleviate salt stress-induced growth inhibition in torenia ‘Kauai Rose’. Plant Biotechnol. Rep. 2023, 17, 203–212.
- Kim, D.; Jeon, S.J.; Yanders, S.; Park, S.C.; Kim, H.S.; Kim, S. MYB3 plays an important role in lignin and anthocyanin biosynthesis under salt stress condition in Arabidopsis. Plant Cell Rep. 2022, 41, 1549–1560.
- Gao, Z.; Ma, C.; Zheng, C.; Yao, Y.; Du, Y. Advances in the regulation of plant salt-stress tolerance by miRNA. Mol. Biol. Rep. 2022, 49, 5041–5055.
- Um, T.; Choi, J.; Park, T.; Chung, P.J.; Jung, S.E.; Shim, J.S.; Kim, Y.S.; Choi, I.Y.; Park, S.C.; Oh, S.J.; et al. Rice microRNA171f/SCL6 module enhances drought tolerance by regulation of flavonoid biosynthesis genes. Plant Direct. 2022, 6, e374.
- Wang, J.; Lian, W.; Cao, Y.; Wang, X.; Wang, G.; Qi, C.; Liu, L.; Qin, S.; Yuan, X.; Li, X.; et al. Overexpression of BoNAC019, a NAC transcription factor from Brassica oleracea, negatively regulates the dehydration response and anthocyanin biosynthesis in Arabidopsis. Sci. Rep. 2018, 8, 13349.
- Jiang, Y.; Wu, X.; Shi, M.; Yu, J.; Guo, C. The miR159-MYB33-ABI5 module regulates seed germination in Arabidopsis. Physiol. Plant. 2022, 174, e13659.
- Wyrzykowska, A.; Bielewicz, D.; Plewka, P.; Soltys-Kalina, D.; Wasilewicz-Flis, I.; Marczewski, W.; Jarmolowski, A.; Szweykowska-Kulinska, Z. The MYB33, MYB65, and MYB101 transcription factors affect Arabidopsis and potato responses to drought by regulating the ABA signaling pathway. Physiol. Plant. 2022, 174, e13775.
- Guang, H.; Xiaoyang, G.; Zhian, W.; Ye, W.; Peng, W.; Linfang, S.; Bingting, W.; Anhong, Z.; Fuguang, L.; Jiahe, W. The cotton MYB33 gene is a hub gene regulating the trade-off between plant growth and defense in Verticillium dahliae infection. J. Adv. Res. 2023; in press.
- Yang, J.; Zhang, N.; Mi, X.; Wu, L.; Ma, R.; Zhu, X.; Yao, L.; Jin, X.; Si, H.; Wang, D. Identification of miR159s and their target genes and expression analysis under drought stress in potato. Comput. Biol. Chem. 2014, 53, 204–213.
- López-Galiano, M.J.; García-Robles, I.; González-Hernández, A.I.; Camañes, G.; Vicedo, B.; Real, M.D.; Rausell, C. Expression of miR159 Is Altered in Tomato Plants Undergoing Drought Stress. Plants 2019, 8, 201.
- Cui, J.; Gao, Z.; Li, B.; Li, J.; Li, X.; Wang, C.; Cheng, D.; Dai, C. Identification of anthocyanin biosynthesis related microRNAs and total microRNAs in Lonicera edulis by high-throughput sequencing. J. Genet. 2020, 99, 31.
- Yang, T.; Wang, Y.; Teotia, S.; Wang, Z.; Shi, C.; Sun, H.; Gu, Y.; Zhang, Z.; Tang, G. The interaction between miR160 and miR165/166 in the control of leaf development and drought tolerance in Arabidopsis. Sci. Rep. 2019, 9, 2832.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.5K
Revisions:
2 times
(View History)
Update Date:
18 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




