Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jianlin Lei | -- | 2602 | 2024-01-31 16:10:26 | | | |
| 2 | Rita Xu | Meta information modification | 2602 | 2024-02-01 03:23:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Lei, J.; Miao, Y.; Bi, W.; Xiang, C.; Li, W.; Zhang, R.; Li, Q.; Yang, Z. Porcine Epidemic Diarrhea Virus. Encyclopedia. Available online: https://encyclopedia.pub/entry/54606 (accessed on 08 February 2026).
Lei J, Miao Y, Bi W, Xiang C, Li W, Zhang R, et al. Porcine Epidemic Diarrhea Virus. Encyclopedia. Available at: https://encyclopedia.pub/entry/54606. Accessed February 08, 2026.
Lei, Jianlin, Yongqiang Miao, Wenrui Bi, Chaohui Xiang, Wei Li, Riteng Zhang, Qian Li, Zengqi Yang. "Porcine Epidemic Diarrhea Virus" Encyclopedia, https://encyclopedia.pub/entry/54606 (accessed February 08, 2026).
Lei, J., Miao, Y., Bi, W., Xiang, C., Li, W., Zhang, R., Li, Q., & Yang, Z. (2024, January 31). Porcine Epidemic Diarrhea Virus. In Encyclopedia. https://encyclopedia.pub/entry/54606
Lei, Jianlin, et al. "Porcine Epidemic Diarrhea Virus." Encyclopedia. Web. 31 January, 2024.
Copy Citation
Porcine epidemic diarrhea virus (PEDV) is a porcine enteric coronavirus, which is one of the main causative agents of porcine epidemic diarrhea (PED), with 100% morbidity and 80–100% mortality in neonatal piglets. Since 2010, large-scale PED caused by highly pathogenic variants of PEDV has occurred successively in China and other countries in the world, posing a great threat to the global pig industry.
porcine epidemic diarrhea virus
enteric coronavirus
highly pathogenic variants
1. Introduction
Coronaviruses (CoVs) have single-stranded, positive-sense RNA viruses with the largest known genomes among all RNA viruses, ranging from 26 to 32 kb, and they are a family of viruses (Alpha-, Beta-, Gamma-, and Deltacoronavirus) that can cause major diseases in humans, such as severe acute respiratory syndrome (SARS), Middle East respiratory syndrome (MERS), and the recent outbreak of SARS-CoV-2 [1][2][3]. In pigs, several different CoVs have been identified, including four Alphacoronaviruses: porcine epidemic diarrhea virus (PEDV), transmissible gastroenteritis virus (TGEV), porcine respiratory coronavirus (PRCV), swine acute diarrhea syndrome-coronavirus (SADS-CoV), one Betacoronavirus: porcine hemagglutinating encephalomyelitis virus (PHEV), and a porcine Deltacoronavirus (PDCoV) [4]. Porcine CoVs have caused significant economic losses to the global pig industry due to their high mortality in neonatal piglets. It is noteworthy that there is the potential for zoonotic transmission because porcine CoVs continue to adapt and evolve in their hosts, changing their tropism. Recently, a study revealed that PDCoVs were identified in Haitian children with acute undifferentiated febrile illness, and the flexibility of the protein and its interaction with the host cell receptor may be impacted by modifications to the spike S1 subunit, which contains the receptor-binding domain [5]. Therefore, the zoonotic transmission of porcine CoVs could pose a threat to human health.
PEDV was originally discovered in the UK in 1971 and then spread to other European countries, where it occasionally broke out in the latter half of the 20th century [6][7]. In China, a comparable case of diarrhea caused by PEDV was initially documented in 1973, and PEDV was discovered in 1984 [8]. Since 2010, a pervasive outbreak of PED has afflicted the southern regions of China, impacting pigs across all age groups. Notably, the PEDV spread rapidly nationwide, and the mortality rate among neonatal piglets approached 100% [9][10]. Following the first reports of PEDV in the United States in 2013, a highly pathogenic PEDV emerged and quickly infected pig populations [11][12]. The virus then quickly spread throughout the pig industry in Europe, North America, and Asia, posing serious economic threats to the pig industry worldwide [13][14][15][16].
2. Etiology
2.1. PEDV Genome and Functions
PEDV is a single-stranded positive-sense RNA virus that belongs to the alphacoronavirus genus. It has a diameter of 95–190 nm, a characteristic nested crown, and a genome size of approximately 28 kb [17]. Seven overlapping open reading frames in the PEDV genome encode the replicase (ORF1a, 1b) nonstructural proteins and the accessory protein ORF3, the spike (S), the envelope (E), the membrane (M), and the nucleocapsid (N) structural proteins [17]. As the major envelope glycoprotein, the S protein promotes the entry of virions into the cell and gives the viral family name its corona-like appearance in electron micrographs. It comprises five domains: a signal peptide; an S1 region that aids in the attachment of virions to cellular receptors; an S2 region that mediates the fusion of the virus with host cells; a transmembrane domain; and a cytoplasmic tail. The S1 domain is further divided into two sub-domains: an N terminal domain and a C terminal domain, which have the potential to be receptor-binding domains [18]. The capacity of the S protein to bind to a receptor and its role in viral entry determine PEDV invasion and release, host range, tissue tropism, cross-species transmission, and trypsin-dependent proliferation [19]. Importantly, the S protein is the major target of induced neutralizing antibodies, and six neutralizing epitopes have been identified: the S10 (aa 19–220) [20], S1A (aa 435–485) [21], COE (aa 499–638) [22], SS2 (aa 748–755), SS6 (aa 764–771) [23], and 2C10 (aa 1368–1374) [24]. In addition, the PEDV S gene is very genetically diverse and is prone to mutation, and it is frequently utilized to evaluate the virulence and genetic variety of the strains that are circulating in the field [25][26]. The M protein, a component of the viral envelope, participates in the assembly and release of viral particles [27]. Furthermore, the recombinant PEDV M protein is used as the antigen in the indirect enzyme-linked immunosorbent assay (ELISA), which has great sensitivity and specificity in identifying PEDV antibodies [28]. Numerous activities in the viral life cycle have been linked to the N protein, such as regulating the production of viral RNA, encasing viral RNA in helical nucleocapsids, and assembling virions [28]. The N protein’s suppression of the host response might account for the increased PEDV replication in Vero E6 cells that overexpress the protein [29]. By delaying the S-phase cell cycle and inhibiting NF-κB from moving to the nucleus during the host cell confronting process, the PEDV N protein suppressed the formation of IFN-λ in IPEC-J2 cells [28][30].
2.2. Emergence of PEDV Strains in China
When the first cases of PED were reported in the UK in 1971, the phrase “epidemic viral diarrhea” (EVD) was coined [31]. Many European countries were impacted by a similar viral diarrhea outbreak in 1976, known as EVD II [32]. One of the EVD samples those Belgian researchers collected in 1978 was identified experimentally as CV777, a new coronavirus strain that causes diarrhea in pigs [6][33]. The disease was collectively named “porcine epidemic diarrhea” (PED) in 1982 [34]. PED reports have been provided on a regular basis in China since 1973. The presence of PEDV in China was not confirmed until 1984 when the causative agent was discovered by utilizing fluorescently labeled antibodies and serum neutralization assays [9]. Between 1984 and the beginning of 2010, PEDV-related epidemics were mostly sporadic or there were local epidemics in some provinces. However, in 2010, a large-scale PED outbreak occurred in southern China [9][10]. Multiple studies have confirmed that emerging PEDV is the culprit of PED in China, and PEDV has mutated compared with previously prevalent strains [9][35][36].
PEDV is classified into genotype I (GI) and GII based on the evolutionary analysis of the S gene (Figure 1). The majority of cell culture-adapted mutant strains generated by continuing passaging in vitro, such as attenuated CV777 and DR13, as well as several classical strains, including the virulent strain CV777 that was first discovered in Europe and Belgium, are classified as PEDV GI [25]. The GI strains have been reported in most of Europe and Asia, they are less virulent than other strains and typically cause sporadic outbreaks [7]. All the PEDVs isolated in China before 2010 belonged to GI, such as PEDV strain JS2008 (GenBank accession No. KC210146) (Figure 1). The main pandemic strain currently circulating in China is GII, which poses an enormous threat to the pig industry in China. In Ohio, USA, a PEDV variant OH851 strain known as PEDV S-INDEL was initially found in 2014 [12]. The S-INDEL strain was later found in China, such as the CH/SCZY44/2017 (GenBank accession No. MH593418) and CH/SCMY/2018 (GenBank accession No. MH061343) strains (Figure 1).
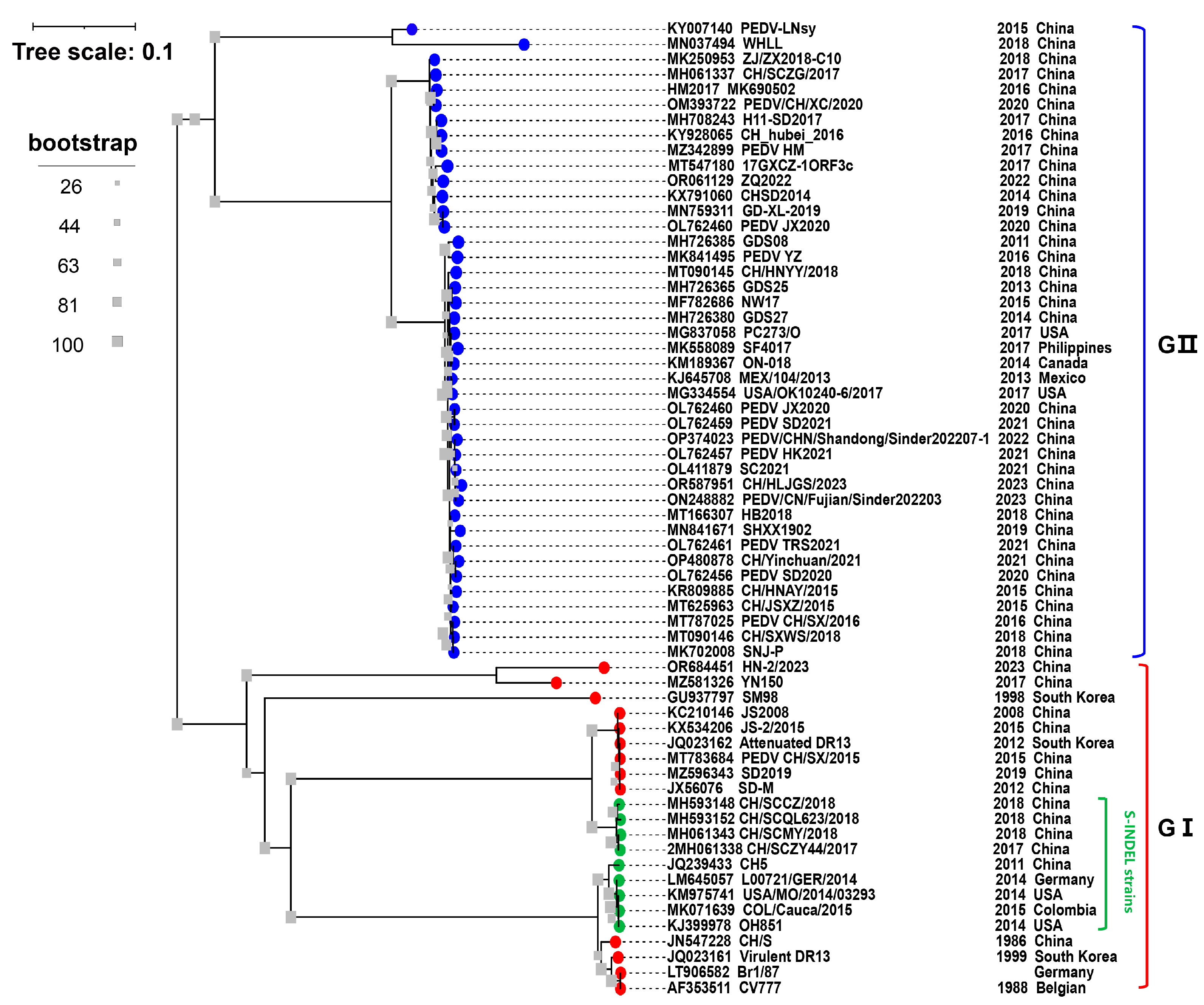
Figure 1. Phylogenetic analysis of the PEDVs based on S gene. The S gene sequences of 64 PEDV strains worldwide were downloaded from GenBank. The tree was constructed using MEGA 7 and visualized using the Interactive Tree Of Life (iTOL) software (version 6.8.1) (accessed on 25 October 2023). The GenBank accession numbers, strain names, collection dates, and countries are shown in the trees. The blue and red dots represent GII and GI, respectively. The green dot represents the PEDV S-INDEL strains. The gray square icon at the branch represents bootstrap values of 1000 replicates.
2.3. PEDV Pathogenicity
PEDV can infect pigs of different ages, and the clinical symptoms are different depending on the virulence of the strain, the immune status of the herd, the age of the affected pigs, and environment variations. The main clinical symptoms of 1- to 7-day-old newborn piglets infected with PEDV were acute watery diarrhea, dehydration, vomiting, and marked emaciation, with 80–100% mortality. Adult pigs infected with PEDV had comparatively low mortality, which was characterized by watery diarrhea, anorexia, agalactia, depression, and impaired reproductive function [37][38].
Early studies confirmed that the classic strain, PEDV CV777 (GI strain), infected piglets 1–20 days old. The clinical symptoms (diarrhea with moderate to severe) appeared at 24–40 h post-inoculation (hpi), and pathological tests also showed marked villus shortening along with the villus height [7]. Since 2010, new non-S-INDEL PEDV strains have emerged, which are highly pathogenic and are known as “highly virulent” PEDV strains based on epidemiologic and clinical data in the field. The emerging non-S-INDEL PEDV strains harbor some novel pathogenic features, such as rapid onset, rapid transmission, rapid death, and high mortality. Additionally, another study reported that the non-S-INDEL PEDV HM2017 strain caused clinical symptoms in piglets at 12 hpi, and they all died at 84 hpi [39]. In the lab, the PEDV CH/Yinchuan/2021 strain infected pig populations of all ages and caused 100% mortality in 3-day-old piglets [38]. Moreover, researchers found that 13-week-old growing pigs infected with PEDV experienced watery diarrhea and vomiting and significantly reduced weight gain despite low mortality (unpublished data). In the challenge test of piglets not separated from their mothers, the sows also showed anorexia after the challenge (unpublished data). Other studies have also confirmed that the highly pathogenic PEDV strains resulted in significant growth retardation in weaned pigs and significant productivity impairment, including reduced sow nursing performance, fewer total pigs and pigs born alive per litter, and lower farrowing rates [7][40]. Strikingly, the pathogenicity may change as PEDV continues to evolve; a new isolate PEDV targets not only the intestinal tract but also the respiratory system in pigs, especially the lungs [41].
2.4. PEDV Transmission
PEDV mainly infects the intestinal tract of pigs, targets small intestinal epithelial cells, and then destroys intestinal villi, resulting in intestinal dysfunction in pigs, causing diarrhea, dehydration, and death, especially for newborn piglets with imperfect digestive systems [42]. Current studies have revealed that PEDV can reach the intestine and cause disease in pigs after entering PEDV-containing media through the mouth and nasal cavity [42][43]. The primary oral–fecal route of PEDV transmission involves direct or indirect contact with pigs that are clinically or subclinically infected as well as diarrheal feces and vomitus [42]. Pigs were infected with PEDV through the consumption of PEDV-contaminated food or exposure to PEDV-contaminated fomites, such as feed, feed ingredients and additives (spray-dried porcine plasma), humans (footwear and clothes), equipment, transport trailers (delivering feed, transporting pigs or carcasses), feed totes, and wild animals (mice, birds, and stray cats) [44][45][46][47][48]. A recent study reported that PEDV colonizing in the intestinal epithelial cells of sows could transfer to CD3+ T cells, which can transmit the virus to the mammary gland through blood circulation and ultimately deliver PEDV to the intestinal tract of piglets through colostrum to cause infection [49]. This finding indicated that sows could transmit PEDV vertically to their offspring through their milk. Additionally, utilizing semen contaminated with PEDV increases the risk of PEDV infection in the pig population [50][51]. Another important route of transmission is the fecal–nasal route, which is basically the airborne transfer of aerosolized PEDV particles into the nose to cause infection in pigs or farms [16][43]. These data indicated that PEDV infected the nasal mucosa, and then the PEDV-loaded dendritic cells transferred the virus to CD3+ T cells and reached the intestine through the blood circulation at the latest, resulting in infection [43].
PEDV transmission in pig populations is influenced by many factors, such as the immune status, biosafety level, and overall health status of pig populations. Feedback feeding is used to control PEDV in some pig farms in China. While this strategy can successfully lower infection, PEDV will recur as time goes on, and PEDV circulation will be formed when the overall antibody level in the pig population is reduced. In addition, although most farms are immunized with vaccines, there is a lack of real-time monitoring of antibody levels in the herd, which leads to the incorrect implementation of vaccine programming and vaccine selection. Comprehensive disinfection is necessary to prevent PED-infected farms since the virus may persist on birthing beds and other associated equipment, possibly causing secondary infections. In fattening pigs and sows, PEDV usually manifests as a subclinical disease with minimal clinical signs (diarrhea). Nevertheless, these herds keep shedding, which facilitates the transmission of PEDV to vulnerable newborn piglets and leads to a high number of fatalities. Therefore, pathogen monitoring and the removal of pathogen-infected pigs are key to the healthy breeding of the system.
3. Epidemiology of PEDV in China
The pig diarrheal disease was first recorded in 1966, it occurs seasonally, usually in the autumn and winter, and it has a localized epidemic pattern [8]. After PEDV was identified in 1984, systematic epidemiological investigations began in China [8]. A general survey of PED epidemics in some provinces, municipalities, and autonomous regions of China revealed that PED was responsible for 1.74% of the overall mortality from 36 pig diseases between 1987 and 1989, whereas TGE was responsible for 9.53% [9]. In 2004, an epidemiological investigation of PEDV in Guangxi Province showed that all pig herds had 42% morbidity and 5.69% mortality, while newborn piglets had 46.4% morbidity and 6.16% mortality, and there was 19.5% morbidity for sows, but no deaths were reported [9]. From 1984 to the beginning of 2010, PEDV was mainly sporadic or regionally endemic in China, with a low mortality rate in piglets [8]. Prior to 2010, PEDVs circulating in China belonged to GⅠ based on full-gene phylogenetic analysis [9]. Importantly, before 2010, the PEDV pandemic was effectively controlled with the widespread use of early-stage Chinese-developed inactivated tissue vaccines, inactivated CV777 vaccines, and inactivated or attenuated bivalent vaccines (PEDV and TGEV) [9].
In the winter of 2010, a large PED outbreak that began in southern China quickly swept throughout the nation, killing millions of piglets and catastrophically harming the pig industry (Figure 2A,B). Of note, the vaccine-immunized farms were not spared, and the mortality rate was close to 100% in neonatal piglets [10][11][35]. It has been confirmed by some investigations that PEDV has mutated in China, and the traditional vaccine PEDV CV777 cannot completely protect against highly pathogenic variants [9][10][38]. These variants have higher virulence and cause clinical symptoms in pigs of all ages, with 80–100% mortality in piglets [37][38]. In 29 Chinese provinces, excluding Tibet and Hainan, an epidemiological survey carried out between February 2011 and March 2014 found that the rate of PEDV-positive samples ranged from 61.10% to 78.49%, while the rate of PEDV-positive pig farms was 71.43% to 83.47% [9]. These data are essentially comparable with those from other countries. In 2013, data showed that piglet mortality was 90–95% and morbidity was almost 100% in the United States [11][52]. In Germany, the mortality rate for piglets infected with PEDV strains was more than 70% as of 2014 [53]. Recent studies have revealed a seropositivity of PEDV in Croatia as high as 82.8% [54]. Zhang et al. collected 149,869 clinical samples of feces and intestinal tissues from pigs from seven provinces and Shanghai City in China from 2011 to 2021 for possible pathogen identification. The results revealed that PEDV was the major causative agent, with a positive rate of more than 40%, while the positive rates of RV and PDCoV were relatively low at 1–20% and 0–14%, respectively. However, 3.21% of the samples were co-infected with PEDV, TGEV, porcine rotavirus (PoRV), PDCoV, or SADS-CoV, and 31.28% of the samples remained undiagnosed [55]. These data indicate that PEDV is the main pathogen causing PED at present, but PoRV and PDCoV cannot be ignored, and other possible pathogens need to be further identified. A recent study revealed that the Guangdong and Henan provinces are hubs for PEDV transmission in China, and the live pig trade may play a major role in disseminating the virus [56]. Since the African swine fever virus (ASFV) invaded China in 2018, the impact of PEDV on the pig industry has been underestimated, but recent statistics have revealed that PEDV is still widespread in the Chinese pig population (http://www.moa.gov.cn/was5/web/search) (accessed on 25 October 2023).
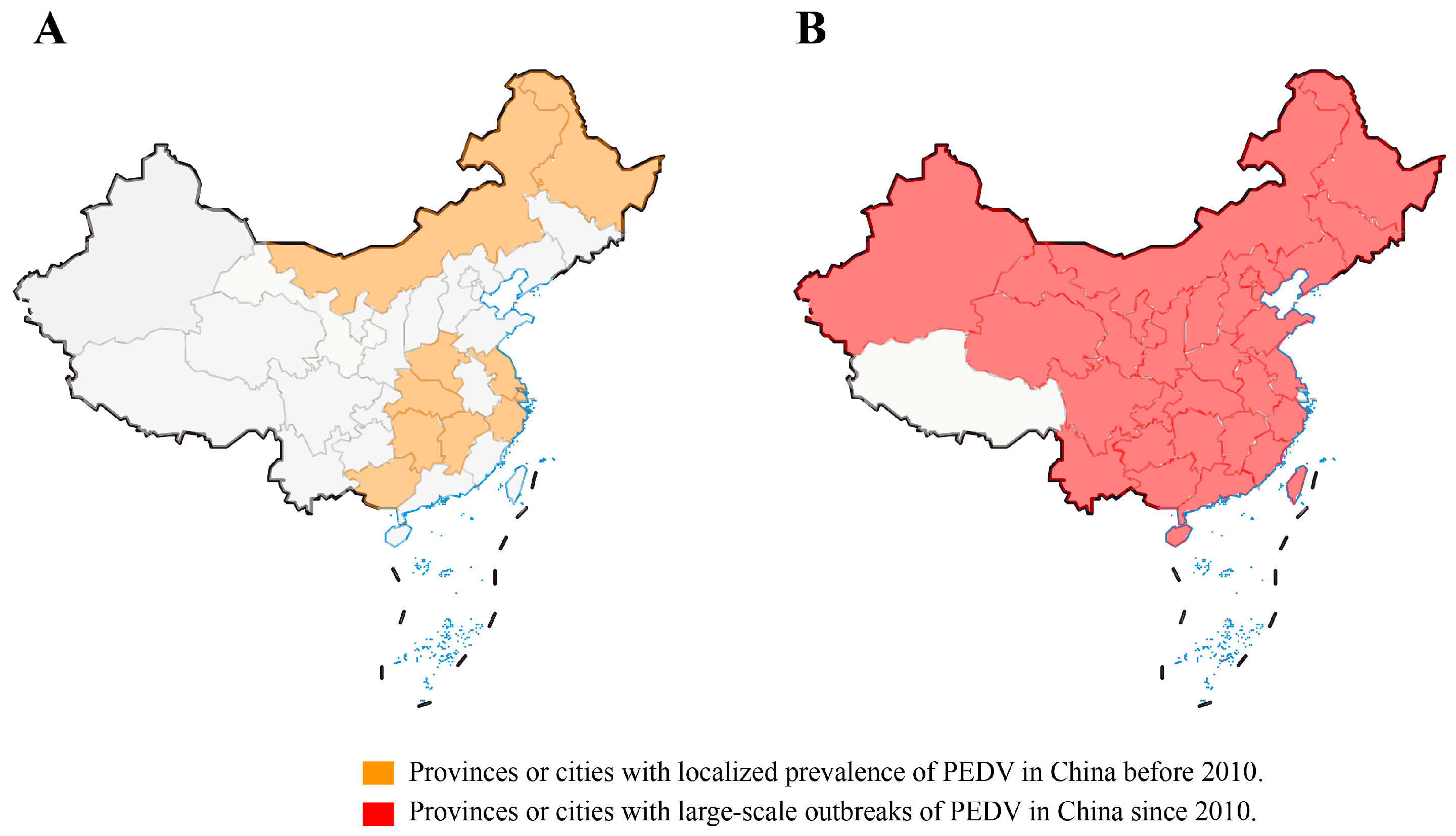
Figure 2. Epidemic distribution of PEDV in provinces and cities in China before and after 2010. (A) Provinces or cities with localized prevalence of PEDV in China before 2010. (B) Provinces or cities with large-scale outbreaks of PEDV in China since 2010.
References
- Drosten, C.; Gunther, S.; Preiser, W.; van der Werf, S.; Brodt, H.R.; Becker, S.; Rabenau, H.; Panning, M.; Kolesnikova, L.; Fouchier, R.A. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003, 348, 1967–1976.
- Zaki, A.M.; van Boheemen, S.; Bestebroer, T.M.; Osterhaus, A.D.; Fouchier, R.A. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med. 2012, 367, 1814–1820.
- Sun, J.; He, W.T.; Wang, L.; Lai, A.; Ji, X.; Zhai, X.; Li, G.; Suchard, M.A.; Tian, J.; Zhou, J.; et al. COVID-19: Epidemiology, evolution, and cross-disciplinary perspectives. Trends Mol. Med. 2020, 26, 483–495.
- Wang, Q.; Vlasova, A.N.; Kenney, S.P.; Saif, L.J. Emerging and re-emerging coronaviruses in pigs. Curr. Opin. Virol. 2019, 34, 39–49.
- Lednicky, J.A.; Tagliamonte, M.S.; White, S.K.; Elbadry, M.A.; Alam, M.M.; Stephenson, C.J.; Bonny, T.S.; Loeb, J.C.; Telisma, T.; Chavannes, S.; et al. Independent infections of porcine deltacoronavirus among Haitian children. Nature 2021, 600, 133–137.
- Pensaert, M.B.; de Bouck, P. A new coronavirus-like particle associated with diarrhea in swine. Arch. Virol. 1978, 58, 243–247.
- Lin, C.M.; Saif, L.J.; Marthaler, D.; Wang, Q. Evolution, antigenicity and pathogenicity of global porcine epidemic diarrhea virus strains. Virus Res. 2016, 226, 20–39.
- Sun, D.; Wang, X.; Wei, S.; Chen, J.; Feng, L. Epidemiology and vaccine of porcine epidemic diarrhea virus in China: A mini-review. J. Vet. Med. Sci. 2016, 78, 355–363.
- Wang, D.; Fang, L.; Xiao, S. Porcine epidemic diarrhea in China. Virus Res. 2016, 226, 7–13.
- Li, W.; Li, H.; Liu, Y.; Pan, Y.; Deng, F.; Song, Y.; Tang, X.; He, Q. New variants of porcine epidemic diarrhea virus, China, 2011. Emerg. Infect. Dis. 2012, 18, 1350–1353.
- Stevenson, G.W.; Hoang, H.; Schwartz, K.J.; Burrough, E.R.; Sun, D.; Madson, D.; Cooper, V.L.; Pillatzki, A.; Gauger, P.; Schmitt, B.J.; et al. Emergence of porcine epidemic diarrhea virus in the United States: Clinical signs, lesions, and viral genomic sequences. J. Vet. Diagn. Invest. 2013, 25, 649–654.
- Wang, L.; Byrum, B.; Zhang, Y. Detection and genetic characterization of deltacoronavirus in pigs, Ohio, USA, 2014. Emerg. Infect. Dis. 2014, 20, 1227–1230.
- Huang, Y.W.; Dickerman, A.W.; Piñeyro, P.; Li, L.; Fang, L.; Kiehne, R.; Opriessnig, T.; Meng, X.J. Origin, evolution, and genotyping of emergent porcine epidemic diarrhea virus strains in the United States. mBio 2013, 4, e00737-13.
- Chen, Q.; Li, G.; Stasko, J.; Thomas, J.T.; Stensland, W.R.; Pillatzki, A.E.; Gauger, P.C.; Schwartz, K.J.; Madson, D.; Yoon, K.J.; et al. Isolation and characterization of porcine epidemic diarrhea viruses associated with the 2013 disease outbreak among swine in the United States. J. Clin. Microbiol. 2014, 52, 234–243.
- Hanke, D.; Pohlmann, A.; Sauter-Louis, C.; Höper, D.; Stadler, J.; Ritzmann, M.; Steinrigl, A.; Schwarz, B.A.; Akimkin, V.; Fux, R.; et al. Porcine epidemic diarrhea in Europe: In-detail analyses of disease dynamics and molecular epidemiology. Viruses 2017, 9, 177.
- Jung, K.; Saif, L.J.; Wang, Q. Porcine epidemic diarrhea virus (PEDV): An update on etiology, transmission, pathogenesis, and prevention and control. Virus Res. 2020, 286, 198045.
- Kocherhans, R.; Bridgen, A.; Ackermann, M.; Tobler, K. Completion of the porcine epidemic diarrhoea coronavirus (PEDV) genome sequence. Virus Genes 2001, 23, 137–144.
- Walls, A.C.; Tortorici, M.A.; Bosch, B.J.; Frenz, B.; Rottier, P.J.M.; DiMaio, F.; Rey, F.A.; Veesler, D. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 2016, 531, 114–117.
- Hu, Y.; Xie, X.; Yang, L.; Wang, A. A comprehensive view on the host factors and viral proteins associated with porcine epidemic diarrhea virus infection. Front. Microbiol. 2021, 12, 762358.
- Li, C.; Li, W.; Lucio de Esesarte, E.; Guo, H.; van den Elzen, P.; Aarts, E.; van den Born, E.; Rottier, P.J.M.; Bosch, B.J. Cell attachment domains of the porcine epidemic diarrhea virus spike protein are key targets of neutralizing antibodies. J. Virol. 2017, 91, e00273-17.
- Chang, C.Y.; Cheng, I.C.; Chang, Y.C.; Tsai, P.S.; Lai, S.Y.; Huang, Y.L.; Jeng, C.R.; Pang, V.F.; Chang, H.W. Identification of neutralizing monoclonal antibodies targeting novel conformational epitopes of the porcine epidemic diarrhoea virus spike protein. Sci. Rep. 2019, 9, 2529.
- Chang, S.H.; Bae, J.L.; Kang, T.J.; Kim, J.; Chung, G.H.; Lim, C.W.; Laude, H.; Yang, M.S.; Jang, Y.S. Identification of the epitope region capable of inducing neutralizing antibodies against the porcine epidemic diarrhea virus. Mol. Cells 2002, 14, 295–299.
- Okda, F.A.; Lawson, S.; Singrey, A.; Nelson, J.; Hain, K.S.; Joshi, L.R.; Christopher-Hennings, J.; Nelson, E.A.; Diel, D.G. The S2 glycoprotein subunit of porcine epidemic diarrhea virus contains immunodominant neutralizing epitopes. Virology 2017, 509, 185–194.
- Cruz, D.J.; Kim, C.J.; Shin, H.J. The GPRLQPY motif located at the carboxy-terminal of the spike protein induces antibodies that neutralize porcine epidemic diarrhea virus. Virus Res. 2008, 132, 192–196.
- Sato, T.; Takeyama, N.; Katsumata, A.; Tuchiya, K.; Kodama, T.; Kusanagi, K. Mutations in the spike gene of porcine epidemic diarrhea virus associated with growth adaptation in vitro and attenuation of virulence in vivo. Virus Genes 2011, 43, 72–78.
- Li, D.; Li, Y.; Liu, Y.; Chen, Y.; Jiao, W.; Feng, H.; Wei, Q.; Wang, J.; Zhang, Y.; Zhang, G. Isolation and identification of a recombinant porcine epidemic diarrhea virus with a novel insertion in S1 domain. Front. Microbiol. 2021, 12, 667084.
- Klumperman, J.; Locker, J.K.; Meijer, A.; Horzinek, M.C.; Geuze, H.J.; Rottier, P.J. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J. Virol. 1994, 68, 6523–6534.
- Fan, J.H.; Zuo, Y.Z.; Shen, X.Q.; Gu, W.Y.; Di, J.M. Development of an enzyme-linked immunosorbent assay for the monitoring and surveillance of antibodies to porcine epidemic diarrhea virus based on a recombinant membrane protein. J. Virol. Methods 2015, 225, 90–94.
- Li, C.; Su, M.; Yin, B.; Guo, D.; Wei, S.; Kong, F.; Feng, L.; Wu, R.; Sun, D. Integrin αvβ3 enhances replication of porcine epidemic diarrhea virus on Vero E6 and porcine intestinal epithelial cells. Vet. Microbiol. 2019, 237, 108400.
- Shan, Y.; Liu, Z.Q.; Li, G.W.; Chen, C.; Luo, H.; Liu, Y.J.; Zhuo, X.H.; Shi, X.F.; Fang, W.H.; Li, X.L. Nucleocapsid protein from porcine epidemic diarrhea virus isolates can antagonize interferon-λ production by blocking the nuclear factor-κB nuclear translocation. J. Zhejiang Univ. Sci. B 2018, 19, 570–580.
- Oldham, J. Letter to the editor. Pig. Farming. 1972, 10, 72–73.
- Chasey, D.; Cartwright, S.F. Virus-like particles associated with porcine epidemic diarrhoea. Res. Vet. Sci. 1978, 25, 255–256.
- Debouck, P.; Pensaert, M. Experimental infection of pigs with a new porcine enteric coronavirus, CV 777. Am. J. Vet. Res. 1980, 41, 219–223.
- Pensaert, M.B.; Callebaut, P.; Debouck, P. Porcine epidemic diarrhea (PED) caused by a coronavirus: Present knowledge. Proc. Congr. Int. Pig. Vet. Soc. 1982, 7, 17–19.
- Wang, X.M.; Niu, B.B.; Yan, H.; Gao, D.S.; Yang, X.; Chen, L.; Chang, H.T.; Zhao, J.; Wang, C.Q. Genetic properties of endemic Chinese porcine epidemic diarrhea virus strains isolated since 2010. Arch. Virol. 2013, 158, 2487–2494.
- Chen, J.; Liu, X.; Shi, D.; Shi, H.; Zhang, X.; Feng, L. Complete genome sequence of a porcine epidemic diarrhea virus variant. J. Virol. 2012, 86, 3408.
- Crawford, K.; Lager, K.; Miller, L.; Opriessnig, T.; Gerber, P.; Hesse, R. Evaluation of porcine epidemic diarrhea virus transmission and the immune response in growing pigs. Vet. Res. 2015, 46, 49.
- Lei, J.L.; Mia, Y.Q.; Guan, Z.; Chen, H.; Xiang, C.H.; Lu, H.Q.; Fang, Y.; Han, Y.; Hu, R.C.; Lu, K.J.; et al. A porcine epidemic diarrhea virus isolated from a sow farm vaccinated with CV777 strain in Yinchuan, China: Characterization, antigenicity, and pathogenicity. Transbound. Emerg. Dis. 2023, 2023, 7082352.
- Yang, D.; Su, M.; Li, C.; Zhang, B.; Qi, S.; Sun, D.; Yin, B. Isolation and characterization of a variant subgroup GII-a porcine epidemic diarrhea virus strain in China. Microb. Pathog. 2020, 140, 103922.
- Goede, D.; Morrison, R.B. Production impact & time to stability in sow herds infected with porcine epidemic diarrhea virus (PEDV). Prev. Vet. Med. 2016, 123, 202–207.
- Van Diep, N.; Choijookhuu, N.; Fuke, N.; Myint, O.; Izzati, U.Z.; Suwanruengsri, M.; Hishikawa, Y.; Yamaguchi, R. New tropisms of porcine epidemic diarrhoea virus (PEDV) in pigs naturally coinfected by variants bearing large deletions in the spike (S) protein and PEDVs possessing an intact S protein. Transbound. Emerg. Dis. 2020, 67, 2589–2601.
- Jung, K.; Saif, L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015, 04, 134–143.
- Li, Y.; Wu, Q.; Huang, L.; Yuan, C.; Wang, J.; Yang, Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018, 9, 3811.
- Pasick, J.; Berhane, Y.; Ojkic, D.; Maxie, G.; Embury-Hyatt, C.; Swekla, K.; Handel, K.; Fairles, J.; Alexandersen, S. Investigation into the role of potentially contaminated feed as a source of the first-detected outbreaks of porcine epidemic diarrhea in Canada. Transbound. Emerg. Dis. 2014, 61, 397–410.
- Dee, S.; Clement, T.; Schelkopf, A.; Nerem, J.; Knudsen, D.; Christopher-Hennings, J.; Nelson, E. An evaluation of contaminated complete feed as a vehicle for porcine epidemic diarrhea virus infection of naïve pigs following consumption via natural feeding behavior: Proof of concept. BMC Vet. Res. 2014, 10, 176.
- Opriessnig, T.; Xiao, C.T.; Gerber, P.F.; Zhang, J.; Halbur, P.G. Porcine epidemic diarrhea virus RNA present in commercial spray-dried porcine plasma is not infectious to naïve pigs. PLoS ONE 2014, 9, e104766.
- Lowe, J.; Gauger, P.; Harmon, K.; Zhang, J.; Connor, J.; Yeske, P.; Loula, T.; Levis, I.; Dufresne, L.; Main, R. Role of transportation in spread of porcine epidemic diarrhea virus infection, United States. Emerg. Infect. Dis. 2014, 20, 872–874.
- Jang, G.; Lee, D.; Shin, S.; Lim, J.; Won, H.; Eo, Y.; Kim, C.H.; Lee, C. Porcine epidemic diarrhea virus: An update overview of virus epidemiology, vaccines, and control strategies in South Korea. J. Vet. Sci. 2023, 24, e58.
- Yuan, C.; Zhang, P.; Liu, P.; Li, Y.; Li, J.; Zhang, E.; Jin, Y.; Yang, Q. A Novel Pathway for Porcine Epidemic Diarrhea Virus Transmission from Sows to Neonatal Piglets Mediated by Colostrum. J. Virol. 2022, 96, e0047722.
- Gallien, S.; Moro, A.; Lediguerher, G.; Catinot, V.; Paboeuf, F.; Bigault, L.; Berri, M.; Gauger, P.C.; Pozzi, N.; Authié, E.; et al. Evidence of porcine epidemic diarrhea virus (PEDV) shedding in semen from infected specific pathogen-free boars. Vet. Res. 2018, 49, 7.
- Gallien, S.; Moro, A.; Lediguerher, G.; Catinot, V.; Paboeuf, F.; Bigault, L.; Gauger, P.C.; Pozzi, N.; Berri, M.; Authié, E.; et al. Limited shedding of an S-InDel strain of porcine epidemic diarrhea virus (PEDV) in semen and questions regarding the infectivity of the detected virus. Vet. Microbiol. 2019, 228, 20–25.
- Cima, G. PED virus reinfecting U.S. herds. Virus estimated to have killed 7 million-plus pigs. J. Am. Vet. Med. Assoc. 2014, 245, 166–167.
- Hanke, D.; Jenckel, M.; Petrov, A.; Ritzmann, M.; Stadler, J.; Akimkin, V.; Blome, S.; Pohlmann, A.; Schirrmeier, H.; Beer, M.; et al. Comparison of porcine epidemic diarrhea viruses from Germany and the United States, 2014. Emerg. Infect. Dis. 2015, 21, 493–496.
- Brnić, D.; Šimić, I.; Lojkić, I.; Krešić, N.; Jungić, A.; Balić, D.; Lolić, M.; Knežević, D.; Hengl, B. The emergence of porcine epidemic diarrhoea in Croatia: Molecular characterization and serology. BMC Vet. Res. 2019, 15, 249.
- Zhang, H.; Zou, C.; Peng, O.; Ashraf, U.; Xu, Q.; Gong, L.; Fan, B.; Zhang, Y.; Xu, Z.; Xue, C.; et al. Global dynamics of porcine enteric coronavirus PEDV epidemiology, evolution, and transmission. Mol. Biol. Evol. 2023, 40, msad052.
- He, W.T.; Bollen, N.; Xu, Y.; Zhao, J.; Dellicour, S.; Yan, Z.; Gong, W.; Zhang, C.; Zhang, L.; Lu, M.; et al. Phylogeography reveals association between swine trade and the spread of porcine epidemic diarrhea virus in China and across the world. Mol. Biol. Evol. 2022, 39, msab364.
More
Information
Subjects:
Veterinary Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
551
Revisions:
2 times
(View History)
Update Date:
01 Feb 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




