Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Attila Seyhan | -- | 3993 | 2024-01-29 16:02:38 | | | |
| 2 | Rita Xu | Meta information modification | 3993 | 2024-01-30 03:09:24 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Seyhan, A.A. miRNA Therapeutics. Encyclopedia. Available online: https://encyclopedia.pub/entry/54491 (accessed on 16 January 2026).
Seyhan AA. miRNA Therapeutics. Encyclopedia. Available at: https://encyclopedia.pub/entry/54491. Accessed January 16, 2026.
Seyhan, Attila A.. "miRNA Therapeutics" Encyclopedia, https://encyclopedia.pub/entry/54491 (accessed January 16, 2026).
Seyhan, A.A. (2024, January 29). miRNA Therapeutics. In Encyclopedia. https://encyclopedia.pub/entry/54491
Seyhan, Attila A.. "miRNA Therapeutics." Encyclopedia. Web. 29 January, 2024.
Copy Citation
The discovery of the link between microRNAs (miRNAs) and a myriad of human diseases, particularly various cancer types, has generated significant interest in exploring their potential as a novel class of drugs. This has led to substantial investments in interdisciplinary research fields such as biology, chemistry, and medical science for the development of miRNA-based therapies. Furthermore, the recent global success of SARS-CoV-2 mRNA vaccines against the COVID-19 pandemic has further revitalized interest in RNA-based immunotherapies, including miRNA-based approaches to cancer treatment. Consequently, RNA therapeutics have emerged as highly adaptable and modular options for cancer therapy.
microRNAs
miRNAs
post-transcriptional gene regulation
miRNA therapeutics
1. Introduction
While most of the research in oncology predominantly centers around the ever-changing aspects of proteins and the RNA molecules responsible for coding those proteins, it is important to note that these coding sequences account for only about 2% of the genome (https://www.genomicseducation.hee.nhs.uk/genotes/knowledge-hub/non-coding-dna/) (accessed on 20 December 2023) [1][2][3]. However, the remaining 98% of the genome, which includes non-coding RNAs (ncRNAs) such as microRNAs (miRNAs), plays pivotal roles in numerous biological processes during normal physiological processes, as well as in the onset and development of various diseases, including various types of human cancers [4]. This emphasizes the significance of miRNAs and other non-coding RNAs in the initiation and progression of tumors.
In addition, miRNAs play key functions in the modulating expression of numerous genes both at the transcriptional [5][6][7] and post-transcriptional [8][9][10][11] levels, and exhibit tissue-specific [12][13] and developmental expression patterns [14][15][16], showcasing their significance in a diverse range of biological processes within cells and organisms. Altered expression of miRNAs has emerged as an additional molecular mechanism implicated in the pathogenesis of numerous diseases [17][18][19], spanning innate immunity [20], autoimmunity and autoimmune diseases [21], viral infections [22][23][24][25], acute hepatitis [26], depression [27], anxiety [28], Alzheimer’s disease [29], Huntington’s disease [30], metabolic and cardiovascular diseases [31][32][33][34], diabetes [8][33][34][35][36][37][38], and many types of cancers [12][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61][62][63][64][65][66][67][68][69]. Consequently, these miRNAs can serve as indicators for the presence of a pathological condition, as well as provide insights into its stage, progression, or genetic associations.
More recently, there is emerging evidence suggesting that diet-derived exogenous miRNAs (or “xenomiRs”) can enter the circulatory system and tissues, potentially influencing gene expression and biological functions [70][71][72][73][74][75].
The absorption of miRNAs by gastric and intestinal cells, along with their potential impact on the gut microbiota and their potential immunomodulatory effects suggests the potential for cross-species or cross-kingdom communication via miRNAs [75]. Because of these observations, one potential method of administering miRNAs is orally.
miRNAs are often associated with extracellular vesicles (EVs), RNA-binding proteins, lipoproteins, or lipid derivatives, along with nanoparticles.
These protective elements shield miRNAs against the harsh gastrointestinal environment, which encompasses salivary and pancreatic RNases, the low pH of the stomach, digestive enzymes, peristaltic activity, and microbial enzymes. This protective shield presumably aids in the absorption of miRNAs from the digestive tract [75].
However, there is ongoing debate surrounding the absorption, stability, and physiological impact of these food-derived miRNAs. Conflicting findings exist regarding the bioavailability and the functional role of plant food-contained miRNAs in human systems [76][77].
Ongoing research continues to uncover new insights into the molecular mechanisms that drive the dysregulation of miRNA biogenesis and aberrant expression in cancer.
For example, it is widely recognized that various factors such as genetic deletions or amplifications, epigenetic methylation of miRNA genomic loci, and modifications affecting the regulation of primary miRNAs (pri-miRNA) by transcription factors, alongside components involved in the miRNA biogenesis pathway frequently lead to alterations in miRNA expression and function across numerous cancer types [56][78][79].
Moreover, other contributing factors, such as oncogenic drivers like mutations occurring in the KRAS gene, also have an impact on the overall miRNA biogenesis and effector function, thereby contributing to broader miRNA dysregulation [80].
As a result, the dysregulation of miRNAs has attracted substantial interest from both academia and industry, standing as a pivotal research domain. This focus extends to comprehending disease biology and exploring their potential applications as diagnostic, prognostic, and predictive biomarkers [68]. Additionally, there is a growing interest in exploring miRNAs as potential drug targets or therapeutic agents [81].
miRNAs are widely recognized as potent genetic regulators that influence diverse biological and developmental processes, while also holding a pivotal role in the pathogenesis of various diseases. This potency stems from a single miRNA’s ability to regulate entire cellular pathways by interacting with numerous target genes [77].
Because of this, miRNAs have emerged as a novel class of therapeutic agents with the potential to restore disrupted cellular functions, particularly in various malignancies, including cancer. However, the very potency of miRNAs can be a double-edged sword. Their far-reaching effects, while beneficial, can also lead to off-target effects in non-targeted tissues, a concern documented in recent clinical trials [82][83][84]. Managing these off-target effects represents a significant challenge to be addressed. Take, for instance, MRX34, a miR-34a mimic encapsulated within a liposome-formulated nanoparticle (NOV40) that was evaluated in a first-in-human phase 1 study in patients with advanced solid tumors, including melanoma NSCLC, hepatocellular carcinoma, and renal carcinoma.
Despite MRX34 demonstrating significant efficacy, with three patients achieving prolonged confirmed partial responses and 14 patients maintaining stable disease (median duration, 136 days) [85], the clinical trial faced termination due to serious immune-mediated adverse events, leading to the deaths of four patients (NCT01829971) [82][83][84]. Nevertheless, the dose-dependent modulation of disease-associated target genes serves as evidence supporting the concept of miRNA-based cancer therapy.
2. miRNAs
Following the discovery of lin-4 as the first miRNA in 1993 in Caenorhabditis elegans [85][86], it became evident that miRNAs are widespread in the animal and plant kingdoms, some of which exhibit high levels of conservation across species [87][88][89].
miRNAs, short non-coding RNA molecules typically about 22 nucleotides long, are naturally encoded in the genomes of diverse species [87][88][89][90].
They play pivotal roles in regulating gene expression at both transcriptional [5][6][7] and post-transcriptional [8][9][10][11][91] levels of their target mRNAs [8][10], influencing mRNA stability and translation [92] across a wide array of biological processes [93], impacting activities such as cell differentiation, proliferation, angiogenesis, and apoptosis.
Additionally, miRNAs demonstrate distinct expression patterns in various tissues [12][13] and during different developmental stages [14][15][16].
There are currently estimated to be more than 2588 mature human miRNAs present in human cells [94], each with a unique temporal and tissue-dependent expression pattern. These miRNAs are estimated to control over 60% of human gene expression, showcasing their significant regulatory roles in diverse physiological processes. Because a single microRNA can regulate multiple genes, many miRNAs can contribute to the development of many human diseases when they become dysfunctional [2][8][18][20][21][22][23][24][25][26][28][30][31][32][33][34][35][37][66][95][96][97] including many types of cancer [39][41][42][43][44][47][50][51][53][55][56][57][58][59][60][61][62][63][64][65][66][67][69][98][99][100][101][102][103].
However, determining the precise relevance of individual miRNAs has been challenging, despite their evident significance as regulatory molecules [104]. Studies investigating miRNA functions through either suppression or overexpression of specific miRNAs have generated data that sometimes conflict with findings from loss-of-function models [104]. For example, studies in Caenorhabditis elegans involving systematic miRNA deletions suggest that fewer than 10% of the miRNAs are individually essential for normal development or viability [105] and this trend appears consistent in mice as well [96].
As illustrated in Figure 1, miRNAs are primarily transcribed from DNA sequences into primary miRNAs (pri-miRNAs), which undergo an initial processing step by Drosha within the nucleus to yield precursor miRNAs (pre-miRNAs) [8][68][106]. It is important to note that up to 40% of miRNA genes might be located within either the introns or exons of other genes [107]. After their transportation from the nucleus to the cytoplasm by exportin 5 (XPO5), pre-miRNAs undergo additional processing by endoribonuclease Dicer, leading to the formation of miRNA duplexes characterized by distinct 3′ overhangs of 2 nucleotides. Subsequently, these miRNA duplexes are loaded onto the Argonaute (AGO) protein, which retains one miRNA strand while discarding the other [10]. The AGO-miRNA complex, along with co-factors like GW182 (TNRC6A), forms the RNA-induced silencing complex (RISC) [91] responsible for cognate mRNA degradation and hence inhibition of translation through interaction with complementary mRNA sequences, typically located within the 3′-untranslated region (3′-UTR) of mRNAs (Figure 1) [108][109][110][111].
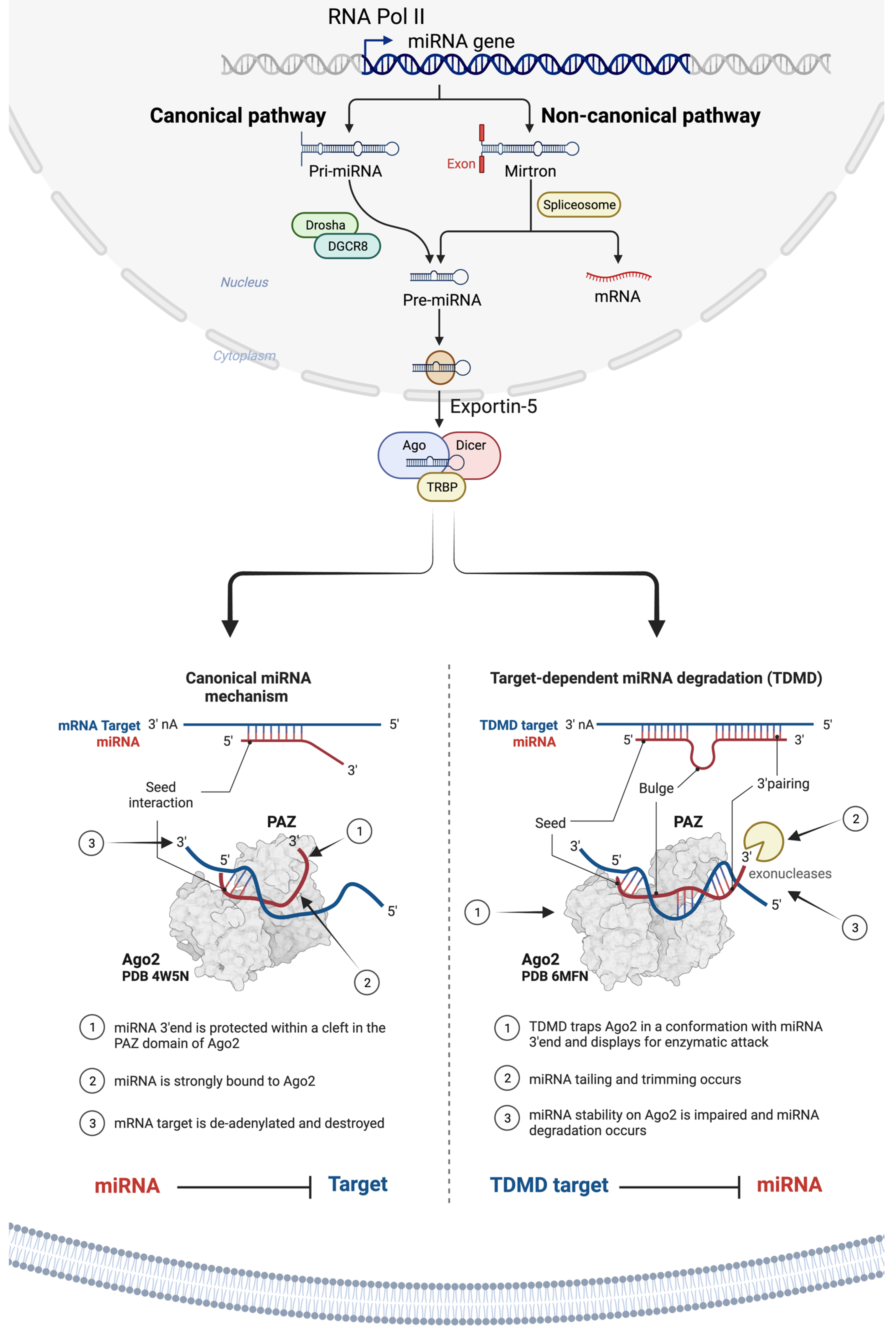
Figure 1. Illustration of miRNA biogenesis, processing, and mechanisms of translational suppression or degradation of target RNA. miRNAs are a class of small, single-stranded non-coding RNAs that function as a guide molecule in RNA silencing and hence modulate the expression of most mRNAs. The miRNA: target–mRNA interaction usually occurs at the 5′ end of the miRNA (i.e., ‘seed’ region). However, recent evidence suggests that there is a special class of target mRNAs which bind the miRNA not only through the ‘seed’ region, but also through a second region of complementarity at the 3′ end of the miRNA. The extended complementarity forces the miRNA out of Ago2, where it becomes accessible to enzymatic degradation. This phenomenon is referred to as the target-directed miRNA degradation mechanism (TDMD).
The interaction between miRNA and target mRNA typically takes place at the 5′ end of the miRNA, referred to as the ‘seed’ region. Yet, recent evidence points to a unique group of target mRNAs that bind the miRNA, not just through the seed but also via a complementary region at the 3′ end of miRNAs. This extended complementarity displaces the miRNA from Ago2, rendering it vulnerable to enzymatic degradation. This process is referred to as the target-directed miRNA degradation mechanism (TDMD) [112][113].
miRNAs are regarded as master regulators of the genome because of their capability to bind to and modify the expression of numerous protein-coding RNAs [114]. Because of this, a single miRNA can potentially regulate distinct mRNAs (anywhere from 10 to 100 protein-coding RNAs) due to their ability to bind to target mRNAs even when the pairing is not perfect [55][115]. As a result, a single miRNA can regulate a range of targets involved in similar cellular processes and pathways, thereby amplifying the cellular response potentially making miRNAs powerful therapeutics to restore perturbed cell functions seen in disease phenotypes. Conversely, a specific messenger RNA can become the target of many miRNAs, whether concurrently or in a context-dependent manner [116], leading to a collaborative repression effect [117][118]. Bioinformatic analyses indicate that a single miRNA can potentially bind to as many as 200 distinct gene targets with various functions, such as transcription factors, receptors, and more (https://bitesizebio.com/24926/mysterious-mirna-identifying-mirnas-and-their-targets/) (accessed on 20 December 2023).
3. miRNAs’ Role in Cancer
Cancer, a complex and heterogeneous disease, is characterized by a sequence of genetic and genomic abnormalities that promote tumorigenesis [119]. These alterations in the genome influence gene function, frequently resulting from genomic aberrations such as chromosomal translocations, amplifications, deletions, insertions, single-nucleotide mutations, or epigenetic modifications. These genetic and epigenetic alterations often result in the activation of oncogenes and the suppression of tumor suppressor genes [120]. In addition, miRNAs have been identified as additional genomic regulators that also play a crucial role in various aspects of organismal development, normal physiological processes, and the development of disease, including many types of cancers [68]. It has been shown that miRNAs play a pivotal in all the known processes involved in cancer, such as proliferation, survival, metastasis, and apoptosis [114]. Data suggest that dysregulation of miRNA function, either through its loss or gain, contributes to cancer development by either upregulating or silencing specific target genes. As a consequence, utilizing miRNAs either as miRNA mimics or antagomirs could present a potent therapeutic strategy to interfere with key molecular pathways associated with cancer as such miRNAs have the capacity to regulate all the recognized hallmarks of cancer, either acting as tumor suppressors or promoting oncogenic processes. Several of these cancer hallmarks influenced by miRNAs are discussed in detail in the literature [65][66].
It is widely accepted that alterations in miRNA genes and their expression are influenced by genetic deletions or amplifications, epigenetic methylation of miRNA gene locations, and modifications affecting pri-miRNA regulation by transcription factors as well as factors involved in miRNA biogenesis, often alter miRNA expression and function across various cancer types [66].
In addition, changes in the miRNA biogenesis process can also impact the availability of target mRNAs, including those associated with the development of cancer [121]. When miRNAs or the machinery involved in miRNA processing are altered or dysregulated this often leads to the loss of normal homeostatic state, leading to malignant transformation, including various types of cancer [51][52][56][65][66][67][122].
Due to their pivotal role in regulating the expression of numerous genes implicated in cellular responses to environmental stressors like hypoxia, oxidative stress, DNA damage, and nutrient deprivation, miRNAs can serve either as oncogenes (oncomirs) or tumor suppressors (onco-suppressor miRs). This is supported by recent findings that have identified miRNAs with oncogenic and tumor-suppressing roles in a range of neoplastic malignancies, and the dysregulation of miRNA expression is closely linked to the initiation, progression, and metastasis of cancer [43][45][103].
Moreover, dysregulated circulating miRNAs have demonstrated associations with disease origin, progression, treatment response, and patient survival [123][124]. For example, the distinctive tissue specificity of miRNAs [13], crucial for the maintenance of normal cells and tissues [40], renders them promising candidates for potential biomarkers in diagnosing cancers of unknown primary [125][126].
Furthermore, with the frequent genetic and epigenetic changes identified in particular miRNAs and the elements involved in miRNA biogenesis across diverse cancer types, oncogenic and tumor suppressor miRNAs have emerged as promising candidates as miRNA-based therapeutics.
4. RNA Therapeutics
As detailed in the literature [127][128], over the past few years, more than 50 siRNA-based drugs have progressed into phase I–III clinical trials. Of those, around 15 programs based on siRNA therapeutics are currently being investigated in phase I, II, and III trials for the treatment of different cancer types [128].
Two siRNA-based drugs, Patisiran and Givosiran (both developed by Alnylam Pharmaceuticals (Cambridge, MA, USA)), obtained approval from the Food and Drug Administration (FDA, Washington, DC, USA) in 2018 and 2019. Patisiran was approved for hereditary transthyretin-mediated amyloidosis, while Givosiran gained approval for acute hepatic porphyria [129][130].
The first example of an FDA-approved RNA-based drug, a siRNA-based therapy developed by Alnylam Pharmaceuticals, is Patisiran, sold under the brand name Onpattro™ for the treatment of polyneuropathy of hereditary transthyretin-mediated amyloidosis in adults. Based on the completion of a successful phase III APOLLO trial, Onpattro™ was approved by the US FDA in August 2018. Onpattro™ contains patisaran, which comprises a siRNA targeting transthyretin (TTR) mRNA conjugated with a lipid complex which leads to a decrease in TTR protein levels in the liver, thus resulting in a reduction in amyloid deposits. Patisiran targets and binds to a genetically conserved sequence found in the 3′UTR of both mutant and wild-type TTR mRNA [131]. Findings from the APOLLO trial, a placebo (77 patients)-controlled phase III trial which enrolled 225 patients showed that 51% of patients receiving Onpattro™ (148 patients, once every three weeks (0.3 mg/kg body weight)) experienced an enhanced quality of life (measured using the Norfolk Quality of Life Diabetic Neuropathy (QoL-DN)), as compared to only 10% of patients in the control group, which received a placebo drug [131][132].
During the COVID-19 pandemic, mRNA technology became instrumental, notably in the development of highly effective mRNA vaccines. These vaccines have played a crucial role in controlling the transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2).
The groundbreaking science behind mRNA vaccines earned Katalin Karikó and Drew Weissman the prestigious 2023 Nobel Prize in Physiology or Medicine for their pioneering work on nucleoside base modifications, enabling the development of these impactful COVID-19 vaccines.
The evolution of cap analogs has vastly improved mRNA translation, while advancements in purification, packaging, and delivery methods have revolutionized medicine. Visionaries like Katalin Karikó, Drew Weissman, Edward Darzynkiewicz, Robert Rhodes, Ugur Sahin, and Ozlem Tureci made pivotal early contributions to mRNA research, deserving recognition for their pioneering efforts. This narrative around mRNA charts a remarkable journey showcasing breakthroughs in a field holding immense promise for the future of medicine.
Consequently, the success of mRNA vaccines has paved the way for integrating mRNA-based technology into personalized neoantigen vaccines, seamlessly incorporating them into the standard oncological workflow [133][134]. These mRNA-based vaccines can be tailored and manufactured as individualized vaccines with multiple neoantigens [135], and can effectively stimulate antigen-presenting cells [136][137][138][139] and be delivered using clinical-stage delivery formulations [140]. The studies and insights from the mRNA-based COVID-19 vaccines highlight the promise of RNA therapeutics as an innovative class of treatments.
However, the effectiveness of miRNA and other nucleic acid-based therapies hinges on a reliable delivery method with minimal adverse events and drug- or treatment-related toxicity.
Delivering miRNA therapeutics to cells poses challenges, requiring precise targeting of diseased cells while sparing healthy ones. In contrast to mRNA COVID-19 vaccines, which are taken up by scavenging immune cells such as dendritic cells and other professional antigen-presenting cells, thus inducing a specific immune response through the processing and expression of the translated mRNA molecules [141], miRNA therapeutics must effectively bypass immune recognition to reach their target cells without triggering an immune response.
5. Advances in the Delivery of miRNA Therapeutics
While a handful of phase 1 and 2 clinical trials have explored miRNA-based therapeutics, there are currently no miRNA-based therapeutics undergoing phase III human clinical trials. This is partly attributed to challenges associated with precisely delivering miRNAs to specific cell types, tissues, and organs.
While several approaches, such as antibodies, nanoparticles, or ligands, have been documented to enhance the effectiveness of miRNAs and decrease off-target effects (like immunotoxicity [142]) when directing miRNAs to specific cells of interest, limitations and challenges persist in the field of miRNA therapeutics.
As illustrated in Figure 2 and discussed in detail in the literature [77][84][143][144], there are various strategies being explored as mechanisms to deliver miRNA therapeutics (mimics and antagomirs) to the indented tissue and to improve pharmacokinetic mechanisms, and avoid off-target effects.
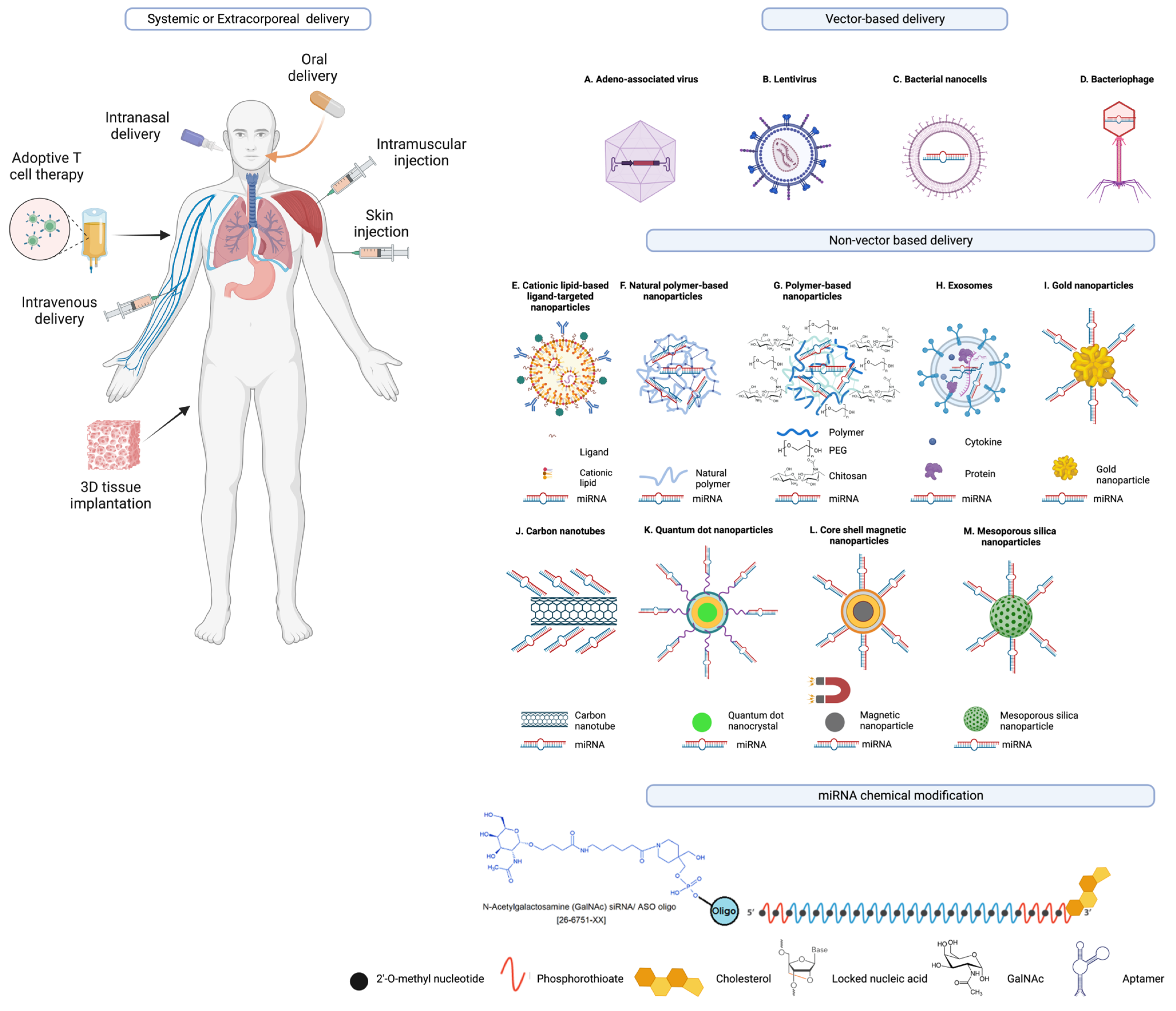
Figure 2. Examples of miRNA delivery systems. miRNA therapeutics can be administered orally or intranasally or through venous (intravenously) or muscle (intramuscularly) or skin (subcutaneously) injections, or via cell-/tissue-directed approaches, or adoptive cell transfer, or the implantation of 3D matrices that release miRNA therapeutics, or other extracorporeal miRNA delivery strategies. Other modes of delivery of miRNA therapeutics include vector based and non-vector-based delivery systems including (A) adeno-associated virus (B) Lentivirus; (C) bacterial nanocells; (D) bacteriophages; liposomes, including monovalent and multivalent lipids such as (E) cationic lipid-based ligand-targeted nanoparticles; (F) natural polymer-based nanoparticles; (G) polymer-based nanoparticles (natural, green and synthetic, blue) conjugated with polyethylene glycol (PEG); (H) extracellular vesicles or exosomes; (I) gold nanoparticles; (J) carbon nanotubes; (K) quantum dot nanoparticles; (L) core–shell magnetic nanoparticles; (M) mesoporous silica nananoparticles and others such as polymeric micelles, and mesoporous silica nanoparticles are the examples of nanocarriers as drug-delivery systems. Moreover, there have been efforts to improve the serum stability, pharmacokinetics, and tissue specificity by targeted delivery of miRNA mimics, miRNA inhibitors, and other nucleic acid therapeutics through the incorporation of various chemical modifications and/or conjugation of these RNA and nucleic acid therapeutics to biomolecules to facilitate receptor-mediated uptake such as N-acetylgalactosamine (GalNAc), 2′-O-methyl nucleotide, phosphorothioate, cholesterol, locked nucleic acid (LNA), and aptamer moieties.
These methods include both vector and non-vector approaches, ranging from lipid-based nanoparticles, polymeric vectors, dendrimer-based vectors, cell-derived membrane vesicles, 3D scaffold-based delivery systems, to various biodegradable and biocompatible nanoparticles derived from polymers and metals [77].
Other strategies for the delivery of RNA-based therapeutics, as discussed in the literature [145], include adeno-associated virus, lentivirus, bacterial nanocells [146], bacteriophages, cationic lipid-based liposomes (including monovalent and multivalent lipids), natural polymer-based nanoparticles, polymer based nanoparticles conjugated with polyethylene glycol (PEG), extracellular vesicles (EVs) or exosomes, nanocomplex-forming functionalized metals such as gold nanoparticles, and carbon nanotubes, polymeric micelles, and mesoporous silica nanoparticles [147], and many others that are engineered to contain biomolecule conjugates for improved stability and pharmacokinetics and target delivery to the intended cell or tissue type [84][144][145][148][149]. A recent study showcased the increased antitumor effectiveness of STING agonists through the covalent attachment of cyclic dinucleotides (CDN) to polymer nanoparticle (poly(β-amino ester) formulation for intravenous delivery [150].
Non-pathogenic recombinant viral vectors, such as retroviruses and lentiviruses (which pose a genomic integration risk), adenoviruses, and adeno-associated viruses (which remain transiently stable in an episomal form within the host cell’s nucleus) [151][152], are under investigation for their capacity to encode the desired RNA transgene. These vectors are being explored for intracellular delivery of miRNA-based therapeutics, making them a significant area of interest [153].
A phase II trial is currently testing an adeno-associated viral vector for the delivery of the miRNA drug AMT-130 for the treatment of Huntington’s disease (ClinicalTrials.gov identifier NCT04120493) [154][155][156]. Despite their potential for the delivery and expression of miRNAs, there are various side effects with the use of viral vectors such as immunogenicity and transgene-related immune responses [157].
The packaging of the negatively charged nucleic acids in liposome nanoparticles masks their negative charge and also protects against serum nuclease degradation [149][158]. Delivery of miRNAs using liposome nanoparticles has already been applied in several clinical studies, such as MRX34 (NCT01829971, NCT02862145) [82][83].
Likewise, bacterial minicells loaded with miRNAs were employed to deliver miR-16 mimics during a phase 1 trial involving patients with recurrent malignant pleural mesothelioma (MesomiR 1, NCT02369198) [159][160]. However, the study also reported several side effects including dose-limiting toxicities, decreased lymphocyte counts, or cardiac events [160].
Extracellular vesicles (EVs), including exosomes, are under exploration as potential drug delivery systems, capable of delivering specific genetic cargo for cellular transfer within the body [161]. For example, EVs derived from mesenchymal stromal cells obtained from human adipose tissue were modified to carry miR-125b. This modification resulted in the inhibition of human hepatocarcinoma cell proliferation [162].
In addition, different modalities of drug delivery systems have been explored for the delivery of miRNA-based drugs such as core–shell magnetic nanoparticles, quantum dot nanocrystals, polymeric micelles, and mesoporous silica nanoparticles are among the other examples of nanocarriers as drug-delivery systems to improve the therapeutic effectiveness and specificity, and tissue targeting of miRNA and other nucleic acid therapeutics [147].
An encouraging strategy involves the covalent conjugation of miRNAs, along with other nucleic acid-based drugs and biomolecules, to lipids, peptides, or sugars. These compounds function through receptor-mediated endocytosis mechanisms [149].
Likewise, a lipophilic cholesterol conjugate was employed to deliver an miR-29-based mimic (remlarsen/MRG-201) to human skin fibroblasts, irrespective of cell type via skin injection in a phase II trial for keloid disorder. The aim was to suppress the expression of extracellular matrix and fibroplasia within the skin (NCT02603224, NCT03601052) [163].
Another approach involves the coupling of aptamer conjugates to specific miRNA therapeutics using a straightforward sticky-end annealing method [164]. This method serves as a strategy for delivering miRNAs to targeted cell types.
Aptamers are single-stranded nucleic acids that are developed as high-affinity ligands specific to cell surface receptors to facilitate the delivery of therapeutic cargo including miRNAs through receptor-mediated transport [149][164]. In ongoing preclinical investigations, researchers are currently investigating aptamer-linked miRNAs, such as the Aptamer-miR-34c conjugate (known as GL21.T-miR-34c) in non-small-cell lung cancer cells [165].
Additional efforts have been made to enhance the serum stability, pharmacokinetics, and tissue specificity of miRNA mimics, miRNA inhibitors, and other nucleic acid therapeutics by incorporating various chemical modifications to miRNA and nucleic acid therapeutics or attaching various biomolecule conjugates to these therapeutic miRNAs to facilitate receptor-mediated uptake such as N-acetylgalactosamine (GalNAc), 2′-O-methyl nucleotide, phosphorothioate, cholesterol, locked nucleic acid (LNA), and aptamer moieties are also shown as examples [77][84][144][166][167].
For example, biomolecule conjugates, such as N-acetylgalactosamine (GalNAc), have been investigated in clinical trials. GalNAc facilitates the targeted delivery of nucleic acid therapeutics through endocytosis by activating liver cell-specific asialoglycoprotein receptors [168][169]. GalNAc linked to a miR-122 inhibitor (RG-101) and a miR-103/107 inhibitor (RG-125/AZD4076) are in clinical trials for chronic HCV [EU Clinical Trials Register (clinicaltrialsregister.eu) EudraCT numbers 2015-001535-21, 2015-004702-42, 2016-002069-77] and steatohepatitis (NCT02612662, NCT02826525), respectively [166][169][170]. However, due to reported side effects such as cases of jaundice, the clinical trial for RG-101 was halted, and investigations into the cause of these effects are ongoing [171][172][173].
Recent preclinical investigations have also explored other examples of GalNAc-conjugated LNA, anti-miR-122 antisense oligonucleotides, or nano-carrier vehicles in combination with cell type-specific biomolecule conjugates or miR-155 inhibitors by gold nanoparticles formulated with antagomir and AS1411 aptamer [174][175].
In addition, the 3D matrices for delivering nucleic acid-based therapeutics and conventional drugs are currently undergoing optimization with diverse design features. This encompasses various application routes, such as edible or injectable carriers [77][176][177][178].
One potential method of administering miRNAs is orally [75]. Data demonstrate that miRNAs are commonly associated with EVs, lipoproteins, or lipid derivatives, and RNA-binding proteins. These associations, along with the use of nanoparticles, shield miRNAs from the harsh conditions in the gastrointestinal tract. This includes protection against salivary and pancreatic RNases, the stomach’s acidic pH, digestive enzymes, peristaltic activity, and microbial enzymes. This safeguarding mechanism is thought to aid in the absorption of miRNAs from the digestive tract [75]. However, there is ongoing debate surrounding the absorption, stability, and physiological impact of these edible or food-derived miRNAs.
References
- Rands, C.M.; Meader, S.; Ponting, C.P.; Lunter, G. 8.2% of the Human genome is constrained: Variation in rates of turnover across functional element classes in the human lineage. PLoS Genet. 2014, 10, e1004525.
- Dunham, I.; Kundaje, A.; Aldred, S.F.; Collins, P.J.; Davis, C.A.; Doyle, F.; Epstein, C.B.; Frietze, S.; Harrow, J.; Kaul, R.; et al. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74.
- Halldorsson, B.V.; Eggertsson, H.P.; Moore, K.H.S.; Hauswedell, H.; Eiriksson, O.; Ulfarsson, M.O.; Palsson, G.; Hardarson, M.T.; Oddsson, A.; Jensson, B.O.; et al. The sequences of 150,119 genomes in the UK Biobank. Nature 2022, 607, 732–740.
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118.
- Khraiwesh, B.; Arif, M.A.; Seumel, G.I.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Transcriptional control of gene expression by microRNAs. Cell 2010, 140, 111–122.
- Samad, A.F.A.; Sajad, M.; Nazaruddin, N.; Fauzi, I.A.; Murad, A.M.A.; Zainal, Z.; Ismail, I. MicroRNA and Transcription Factor: Key Players in Plant Regulatory Network. Front. Plant Sci. 2017, 8, 565.
- Tong, Z.; Cui, Q.; Wang, J.; Zhou, Y. TransmiR v2.0: An updated transcription factor-microRNA regulation database. Nucleic Acids Res. 2018, 47, D253–D258.
- Seyhan, A.A. microRNAs with different functions and roles in disease development and as potential biomarkers of diabetes: Progress and challenges. Mol. Biosyst. 2015, 11, 1217–1234.
- Chen, P.Y.; Meister, G. microRNA-guided posttranscriptional gene regulation. Biol. Chem. 2005, 386, 1205–1218.
- Catalanotto, C.; Cogoni, C.; Zardo, G. MicroRNA in Control of Gene Expression: An Overview of Nuclear Functions. Int. J. Mol. Sci. 2016, 17, 1712.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Wang, H.; Meng, Q.; Qian, J.; Li, M.; Gu, C.; Yang, Y. Review: RNA-based diagnostic markers discovery and therapeutic targets development in cancer. Pharmacol. Ther. 2022, 234, 108123.
- Guo, Z.; Maki, M.; Ding, R.; Yang, Y.; Zhang, B.; Xiong, L. Genome-wide survey of tissue-specific microRNA and transcription factor regulatory networks in 12 tissues. Sci. Rep. 2014, 4, 5150.
- Wienholds, E.; Kloosterman, W.P.; Miska, E.; Alvarez-Saavedra, E.; Berezikov, E.; de Bruijn, E.; Horvitz, H.R.; Kauppinen, S.; Plasterk, R.H. MicroRNA expression in zebrafish embryonic development. Science 2005, 309, 310–311.
- Aboobaker, A.A.; Tomancak, P.; Patel, N.; Rubin, G.M.; Lai, E.C. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl. Acad. Sci. USA 2005, 102, 18017–18022.
- Walker, J.C.; Harland, R.M. Expression of microRNAs during embryonic development of Xenopus tropicalis. Gene Expr. Patterns 2008, 8, 452–456.
- Li, Y.; Kowdley, K.V. MicroRNAs in Common Human Diseases. Genom. Proteom. Bioinform. 2012, 10, 246–253.
- De Guire, V.; Robitaille, R.; Tetreault, N.; Guerin, R.; Menard, C.; Bambace, N.; Sapieha, P. Circulating miRNAs as sensitive and specific biomarkers for the diagnosis and monitoring of human diseases: Promises and challenges. Clin. Biochem. 2013, 46, 846–860.
- Condrat, C.E.; Thompson, D.C.; Barbu, M.G.; Bugnar, O.L.; Boboc, A.; Cretoiu, D.; Suciu, N.; Cretoiu, S.M.; Voinea, S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells 2020, 9, 276.
- Gareev, I.; de Jesus Encarnacion Ramirez, M.; Goncharov, E.; Ivliev, D.; Shumadalova, A.; Ilyasova, T.; Wang, C. MiRNAs and lncRNAs in the regulation of innate immune signaling. Noncoding RNA Res. 2023, 8, 534–541.
- Pauley, K.M.; Cha, S.; Chan, E.K. MicroRNA in autoimmunity and autoimmune diseases. J. Autoimmun. 2009, 32, 189–194.
- Sullivan, C.S.; Ganem, D. MicroRNAs and viral infection. Mol. Cell 2005, 20, 3–7.
- Skalsky, R.L.; Cullen, B.R. Viruses, microRNAs, and host interactions. Annu. Rev. Microbiol. 2010, 64, 123–141.
- Barbu, M.G.; Condrat, C.E.; Thompson, D.C.; Bugnar, O.L.; Cretoiu, D.; Toader, O.D.; Suciu, N.; Voinea, S.C. MicroRNA Involvement in Signaling Pathways During Viral Infection. Front. Cell Dev. Biol. 2020, 8, 143.
- Abu-Izneid, T.; AlHajri, N.; Ibrahim, A.M.; Javed, M.N.; Salem, K.M.; Pottoo, F.H.; Kamal, M.A. Micro-RNAs in the regulation of immune response against SARS CoV-2 and other viral infections. J. Adv. Res. 2021, 30, 133–145.
- Elfimova, N.; Schlattjan, M.; Sowa, J.P.; Dienes, H.P.; Canbay, A.; Odenthal, M. Circulating microRNAs: Promising candidates serving as novel biomarkers of acute hepatitis. Front. Physiol. 2012, 3, 476.
- Li, Y.J.; Xu, M.; Gao, Z.H.; Wang, Y.Q.; Yue, Z.; Zhang, Y.X.; Li, X.X.; Zhang, C.; Xie, S.Y.; Wang, P.Y. Alterations of serum levels of BDNF-related miRNAs in patients with depression. PLoS ONE 2013, 8, e63648.
- Scott, K.A.; Hoban, A.E.; Clarke, G.; Moloney, G.M.; Dinan, T.G.; Cryan, J.F. Thinking small: Towards microRNA-based therapeutics for anxiety disorders. Expert. Opin. Investig. Drugs 2015, 24, 529–542.
- Liu, W.; Liu, C.; Zhu, J.; Shu, P.; Yin, B.; Gong, Y.; Qiang, B.; Yuan, J.; Peng, X. MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer’s-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 2012, 33, 522–534.
- Weir, D.W.; Sturrock, A.; Leavitt, B.R. Development of biomarkers for Huntington’s disease. Lancet Neurol. 2011, 10, 573–590.
- Wang, R.; Li, N.; Zhang, Y.; Ran, Y.; Pu, J. Circulating microRNAs are promising novel biomarkers of acute myocardial infarction. Intern. Med. 2011, 50, 1789–1795.
- Recchioni, R.; Marcheselli, F.; Olivieri, F.; Ricci, S.; Procopio, A.D.; Antonicelli, R. Conventional and novel diagnostic biomarkers of acute myocardial infarction: A promising role for circulating microRNAs. Biomarkers 2013, 18, 547–558.
- Nunez Lopez, Y.O.; Coen, P.M.; Goodpaster, B.H.; Seyhan, A.A. Gastric bypass surgery with exercise alters plasma microRNAs that predict improvements in cardiometabolic risk. Int. J. Obes. 2017, 41, 1121–1130.
- Nunez Lopez, Y.O.; Garufi, G.; Pasarica, M.; Seyhan, A.A. Elevated and Correlated Expressions of miR-24, miR-30d, miR-146a, and SFRP-4 in Human Abdominal Adipose Tissue Play a Role in Adiposity and Insulin Resistance. Int. J. Endocrinol. 2018, 2018, 7351902.
- Wang, Q.; Wang, Y.; Minto, A.W.; Wang, J.; Shi, Q.; Li, X.; Quigg, R.J. MicroRNA-377 is up-regulated and can lead to increased fibronectin production in diabetic nephropathy. FASEB J. 2008, 22, 4126–4135.
- Seyhan, A.A.; Nunez Lopez, Y.O.; Xie, H.; Yi, F.; Mathews, C.; Pasarica, M.; Pratley, R.E. Pancreas-enriched miRNAs are altered in the circulation of subjects with diabetes: A pilot cross-sectional study. Sci. Rep. 2016, 6, 31479.
- Nunez Lopez, Y.O.; Garufi, G.; Seyhan, A.A. Altered levels of circulating cytokines and microRNAs in lean and obese individuals with prediabetes and type 2 diabetes. Mol. Biosyst. 2016, 13, 106–121.
- Nunez Lopez, Y.O.; Pittas, A.G.; Pratley, R.E.; Seyhan, A.A. Circulating levels of miR-7, miR-152 and miR-192 respond to vitamin D supplementation in adults with prediabetes and correlate with improvements in glycemic control. J. Nutr. Biochem. 2017, 49, 117–122.
- Kluiver, J.; Poppema, S.; de Jong, D.; Blokzijl, T.; Harms, G.; Jacobs, S.; Kroesen, B.J.; van den Berg, A. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J. Pathol. 2005, 207, 243–249.
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA expression profiles classify human cancers. Nature 2005, 435, 834–838.
- Esquela-Kerscher, A.; Slack, F.J. Oncomirs—microRNAs with a role in cancer. Nat. Rev. Cancer 2006, 6, 259–269.
- Lawler, S.; Chiocca, E.A. Emerging functions of microRNAs in glioblastoma. J. Neurooncol. 2009, 92, 297–306.
- Ventura, A.; Jacks, T. MicroRNAs and cancer: Short RNAs go a long way. Cell 2009, 136, 586–591.
- Suzuki, H.; Maruyama, R.; Yamamoto, E.; Kai, M. Epigenetic alteration and microRNA dysregulation in cancer. Front. Genet. 2013, 4, 258.
- Di Leva, G.; Garofalo, M.; Croce, C.M. MicroRNAs in cancer. Annu. Rev. Pathol. 2014, 9, 287–314.
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469.
- Graveel, C.R.; Calderone, H.M.; Westerhuis, J.J.; Winn, M.E.; Sempere, L.F. Critical analysis of the potential for microRNA biomarkers in breast cancer management. Breast Cancer Dove Med. Press 2015, 7, 59–79.
- Hata, A.; Lieberman, J. Dysregulation of microRNA biogenesis and gene silencing in cancer. Sci. Signal 2015, 8, re3.
- Lin, S.; Gregory, R.I. MicroRNA biogenesis pathways in cancer. Nat. Rev. Cancer 2015, 15, 321–333.
- Hata, A.; Kashima, R. Dysregulation of microRNA biogenesis machinery in cancer. Crit. Rev. Biochem. Mol. Biol. 2016, 51, 121–134.
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004.
- Lulla, A.R.; Slifker, M.J.; Zhou, Y.; Lev, A.; Einarson, M.B.; Dicker, D.T.; El-Deiry, W.S. miR-6883 Family miRNAs Target CDK4/6 to Induce G(1) Phase Cell-Cycle Arrest in Colon Cancer Cells. Cancer Res. 2017, 77, 6902–6913.
- Biswas, S. MicroRNAs as Therapeutic Agents: The Future of the Battle Against Cancer. Curr. Top. Med. Chem. 2018, 18, 2544–2554.
- Hu, W.; Tan, C.; He, Y.; Zhang, G.; Xu, Y.; Tang, J. Functional miRNAs in breast cancer drug resistance. Onco Targets Ther. 2018, 11, 1529–1541.
- Plotnikova, O.; Baranova, A.; Skoblov, M. Comprehensive Analysis of Human microRNA-mRNA Interactome. Front. Genet. 2019, 10, 933.
- Ali Syeda, Z.; Langden, S.S.S.; Munkhzul, C.; Lee, M.; Song, S.J. Regulatory Mechanism of MicroRNA Expression in Cancer. Int. J. Mol. Sci. 2020, 21, 1723.
- Annese, T.; Tamma, R.; De Giorgis, M.; Ribatti, D. microRNAs Biogenesis, Functions and Role in Tumor Angiogenesis. Front. Oncol. 2020, 10, 581007.
- Cao, D.; Di, M.; Liang, J.; Shi, S.; Tan, Q.; Wang, Z. MicroRNA-183 in Cancer Progression. J. Cancer 2020, 11, 1315–1324.
- Fathi, M.; Ghafouri-Fard, S.; Abak, A.; Taheri, M. Emerging roles of miRNAs in the development of pancreatic cancer. Biomed. Pharmacother. 2021, 141, 111914.
- Galka-Marciniak, P.; Urbanek-Trzeciak, M.O.; Nawrocka, P.M.; Kozlowski, P. A pan-cancer atlas of somatic mutations in miRNA biogenesis genes. Nucleic Acids Res. 2021, 49, 601–620.
- Inoue, J.; Inazawa, J. Cancer-associated miRNAs and their therapeutic potential. J. Hum. Genet. 2021, 66, 937–945.
- Pajares, M.J.; Alemany-Cosme, E.; Goni, S.; Bandres, E.; Palanca-Ballester, C.; Sandoval, J. Epigenetic Regulation of microRNAs in Cancer: Shortening the Distance from Bench to Bedside. Int. J. Mol. Sci. 2021, 22, 7350.
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Hogendorf, P. The Role of microRNA in Pancreatic Cancer. Biomedicines 2021, 9, 1322.
- Smolarz, B.; Durczyński, A.; Romanowicz, H.; Szyłło, K.; Hogendorf, P. miRNAs in Cancer (Review of Literature). Int. J. Mol. Sci. 2022, 23, 2805.
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502.
- Raufi, A.G.; May, M.S.; Hadfield, M.J.; Seyhan, A.A.; El-Deiry, W.S. Advances in Liquid Biopsy Technology and Implications for Pancreatic Cancer. Int. J. Mol. Sci. 2023, 24, 4238.
- Ricarte-Filho, J.C.; Casado-Medrano, V.; Reichenberger, E.; Spangler, Z.; Scheerer, M.; Isaza, A.; Baran, J.; Patel, T.; MacFarland, S.P.; Brodeur, G.M.; et al. DICER1 RNase IIIb domain mutations trigger widespread miRNA dysregulation and MAPK activation in pediatric thyroid cancer. Front. Endocrinol. 2023, 14, 1083382.
- Seyhan, A.A. Circulating microRNAs as Potential Biomarkers in Pancreatic Cancer-Advances and Challenges. Int. J. Mol. Sci. 2023, 24, 13340.
- Abdelaal, A.M.; Sohal, I.S.; Iyer, S.; Sudarshan, K.; Kothandaraman, H.; Lanman, N.A.; Low, P.S.; Kasinski, A.L. A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy. Oncogene 2023, 42, 2985–2999.
- Zhang, L.; Hou, D.; Chen, X.; Li, D.; Zhu, L.; Zhang, Y.; Li, J.; Bian, Z.; Liang, X.; Cai, X.; et al. Exogenous plant MIR168a specifically targets mammalian LDLRAP1: Evidence of cross-kingdom regulation by microRNA. Cell Res. 2012, 22, 107–126.
- Witwer, K.W. XenomiRs and miRNA homeostasis in health and disease: Evidence that diet and dietary miRNAs directly and indirectly influence circulating miRNA profiles. RNA Biol. 2012, 9, 1147–1154.
- Wagner, A.E.; Piegholdt, S.; Ferraro, M.; Pallauf, K.; Rimbach, G. Food derived microRNAs. Food Funct. 2015, 6, 714–718.
- Zhang, L.; Chen, T.; Yin, Y.; Zhang, C.Y.; Zhang, Y.L. Dietary microRNA-A Novel Functional Component of Food. Adv. Nutr. 2019, 10, 711–721.
- Díez-Sainz, E.; Lorente-Cebrián, S.; Aranaz, P.; Riezu-Boj, J.I.; Martínez, J.A.; Milagro, F.I. Potential Mechanisms Linking Food-Derived MicroRNAs, Gut Microbiota and Intestinal Barrier Functions in the Context of Nutrition and Human Health. Front. Nutr. 2021, 8, 586564.
- Cieślik, M.; Bryniarski, K.; Nazimek, K. Dietary and orally-delivered miRNAs: Are they functional and ready to modulate immunity? AIMS Allergy Immunol. 2023, 7, 104–131.
- Dickinson, B.; Zhang, Y.; Petrick, J.S.; Heck, G.; Ivashuta, S.; Marshall, W.S. Lack of detectable oral bioavailability of plant microRNAs after feeding in mice. Nat. Biotechnol. 2013, 31, 965–967.
- Diener, C.; Keller, A.; Meese, E. Emerging concepts of miRNA therapeutics: From cells to clinic. Trends Genet. 2022, 38, 613–626.
- Arif, K.M.T.; Elliott, E.K.; Haupt, L.M.; Griffiths, L.R. Regulatory Mechanisms of Epigenetic miRNA Relationships in Human Cancer and Potential as Therapeutic Targets. Cancers 2020, 12, 2922.
- Machowska, M.; Galka-Marciniak, P.; Kozlowski, P. Consequences of genetic variants in miRNA genes. Comput. Struct. Biotechnol. J. 2022, 20, 6443–6457.
- Bortoletto, A.S.; Parchem, R.J. KRAS Hijacks the miRNA Regulatory Pathway in Cancer. Cancer Res. 2023, 83, 1563–1572.
- Li, L.J.; Leng, R.X.; Fan, Y.G.; Pan, H.F.; Ye, D.Q. Translation of noncoding RNAs: Focus on lncRNAs, pri-miRNAs, and circRNAs. Exp. Cell Res. 2017, 361, 1–8.
- Beg, M.S.; Brenner, A.J.; Sachdev, J.; Borad, M.; Kang, Y.K.; Stoudemire, J.; Smith, S.; Bader, A.G.; Kim, S.; Hong, D.S. Phase I study of MRX34, a liposomal miR-34a mimic, administered twice weekly in patients with advanced solid tumors. Invest. New Drugs 2017, 35, 180–188.
- Hong, D.S.; Kang, Y.K.; Borad, M.; Sachdev, J.; Ejadi, S.; Lim, H.Y.; Brenner, A.J.; Park, K.; Lee, J.L.; Kim, T.Y.; et al. Phase 1 study of MRX34, a liposomal miR-34a mimic, in patients with advanced solid tumours. Br. J. Cancer 2020, 122, 1630–1637.
- Desantis, V.; Saltarella, I.; Lamanuzzi, A.; Melaccio, A.; Solimando, A.G.; Mariggiò, M.A.; Racanelli, V.; Paradiso, A.; Vacca, A.; Frassanito, M.A. MicroRNAs-Based Nano-Strategies as New Therapeutic Approach in Multiple Myeloma to Overcome Disease Progression and Drug Resistance. Int. J. Mol. Sci. 2020, 21, 3084.
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854.
- Wightman, B.; Ha, I.; Ruvkun, G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell 1993, 75, 855–862.
- Pasquinelli, A.E.; Reinhart, B.J.; Slack, F.; Martindale, M.Q.; Kuroda, M.I.; Maller, B.; Hayward, D.C.; Ball, E.E.; Degnan, B.; Müller, P.; et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature 2000, 408, 86–89.
- Li, S.C.; Chan, W.C.; Hu, L.Y.; Lai, C.H.; Hsu, C.N.; Lin, W.C. Identification of homologous microRNAs in 56 animal genomes. Genomics 2010, 96, 1–9.
- Friedländer, M.R.; Lizano, E.; Houben, A.J.S.; Bezdan, D.; Báñez-Coronel, M.; Kudla, G.; Mateu-Huertas, E.; Kagerbauer, B.; González, J.; Chen, K.C.; et al. Evidence for the biogenesis of more than 1000 novel human microRNAs. Genome Biol. 2014, 15, R57.
- Griffiths-Jones, S.; Grocock, R.J.; van Dongen, S.; Bateman, A.; Enright, A.J. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006, 34, D140–D144.
- Pu, M.; Chen, J.; Tao, Z.; Miao, L.; Qi, X.; Wang, Y.; Ren, J. Regulatory network of miRNA on its target: Coordination between transcriptional and post-transcriptional regulation of gene expression. Cell. Mol. Life Sci. 2019, 76, 441–451.
- van Rooij, E. The art of microRNA research. Circ. Res. 2011, 108, 219–234.
- Friedman, R.C.; Farh, K.K.; Burge, C.B.; Bartel, D.P. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009, 19, 92–105.
- de Rie, D.; Abugessaisa, I.; Alam, T.; Arner, E.; Arner, P.; Ashoor, H.; Åström, G.; Babina, M.; Bertin, N.; Burroughs, A.M.; et al. An integrated expression atlas of miRNAs and their promoters in human and mouse. Nat. Biotechnol. 2017, 35, 872–878.
- Gambardella, S.; Rinaldi, F.; Lepore, S.M.; Viola, A.; Loro, E.; Angelini, C.; Vergani, L.; Novelli, G.; Botta, A. Overexpression of microRNA-206 in the skeletal muscle from myotonic dystrophy type 1 patients. J. Transl. Med. 2010, 8, 48.
- Baer, C.; Claus, R.; Frenzel, L.P.; Zucknick, M.; Park, Y.J.; Gu, L.; Weichenhan, D.; Fischer, M.; Pallasch, C.P.; Herpel, E.; et al. Extensive promoter DNA hypermethylation and hypomethylation is associated with aberrant microRNA expression in chronic lymphocytic leukemia. Cancer Res. 2012, 72, 3775–3785.
- Achey, R.L.; Khanna, V.; Ostrom, Q.T.; Kruchko, C.; Barnholtz-Sloan, J.S. Incidence and survival trends in oligodendrogliomas and anaplastic oligodendrogliomas in the United States from 2000 to 2013: A CBTRUS Report. J. Neurooncol. 2017, 133, 17–25.
- Eder, M.; Scherr, M. MicroRNA and lung cancer. N. Engl. J. Med. 2005, 352, 2446–2448.
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477.
- Allen, J.E.; Crowder, R.N.; El-Deiry, W.S. First-In-Class Small Molecule ONC201 Induces DR5 and Cell Death in Tumor but Not Normal Cells to Provide a Wide Therapeutic Index as an Anti-Cancer Agent. PLoS ONE 2015, 10, e0143082.
- Bedewy, A.M.L.; Elmaghraby, S.M.; Shehata, A.A.; Kandil, N.S. Prognostic Value of miRNA-155 Expression in B-Cell Non-Hodgkin Lymphoma. Turk. J. Haematol. 2017, 34, 207–212.
- Kline, C.L.B.; Ralff, M.D.; Lulla, A.R.; Wagner, J.M.; Abbosh, P.H.; Dicker, D.T.; Allen, J.E.; El-Deiry, W.S. Role of Dopamine Receptors in the Anticancer Activity of ONC201. Neoplasia 2018, 20, 80–91.
- Lu, F.; Zhao, X.; Zhang, Z.; Xiong, M.; Wang, Y.; Sun, Y.; He, B.; Zhu, J. The diagnostic and prognostic value of the miR-17-92 cluster in hepatocellular carcinoma: A meta-analysis. Front. Genet. 2022, 13, 927079.
- Vidigal, J.A.; Ventura, A. The biological functions of miRNAs: Lessons from in vivo studies. Trends Cell Biol. 2015, 25, 137–147.
- Lujambio, A.; Ropero, S.; Ballestar, E.; Fraga, M.F.; Cerrato, C.; Setien, F.; Casado, S.; Suarez-Gauthier, A.; Sanchez-Cespedes, M.; Git, A.; et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res. 2007, 67, 1424–1429.
- O’Carroll, D.; Schaefer, A. General principals of miRNA biogenesis and regulation in the brain. Neuropsychopharmacology 2013, 38, 39–54.
- Rodriguez, A.; Griffiths-Jones, S.; Ashurst, J.L.; Bradley, A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004, 14, 1902–1910.
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233.
- Iwasaki, S.; Kobayashi, M.; Yoda, M.; Sakaguchi, Y.; Katsuma, S.; Suzuki, T.; Tomari, Y. Hsc70/Hsp90 chaperone machinery mediates ATP-dependent RISC loading of small RNA duplexes. Mol. Cell 2010, 39, 292–299.
- Shukla, G.C.; Singh, J.; Barik, S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol. Cell Pharmacol. 2011, 3, 83–92.
- Akgul, B.; Erdogan, I. Intracytoplasmic Re-localization of miRISC Complexes. Front. Genet. 2018, 9, 403.
- Ghini, F.; Rubolino, C.; Climent, M.; Simeone, I.; Marzi, M.J.; Nicassio, F. Endogenous transcripts control miRNA levels and activity in mammalian cells by target-directed miRNA degradation. Nat. Commun. 2018, 9, 3119.
- Sheu-Gruttadauria, J.; Pawlica, P.; Klum, S.M.; Wang, S.; Yario, T.A.; Schirle Oakdale, N.T.; Steitz, J.A.; MacRae, I.J. Structural Basis for Target-Directed MicroRNA Degradation. Mol. Cell 2019, 75, 1243–1255.e7.
- Garofalo, M.; Croce, C.M. microRNAs: Master regulators as potential therapeutics in cancer. Annu. Rev. Pharmacol. Toxicol. 2011, 51, 25–43.
- Selbach, M.; Schwanhäusser, B.; Thierfelder, N.; Fang, Z.; Khanin, R.; Rajewsky, N. Widespread changes in protein synthesis induced by microRNAs. Nature 2008, 455, 58–63.
- Uhlmann, S.; Mannsperger, H.; Zhang, J.D.; Horvat, E.; Schmidt, C.; Küblbeck, M.; Henjes, F.; Ward, A.; Tschulena, U.; Zweig, K.; et al. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012, 8, 570.
- Grimson, A.; Farh, K.K.; Johnston, W.K.; Garrett-Engele, P.; Lim, L.P.; Bartel, D.P. MicroRNA targeting specificity in mammals: Determinants beyond seed pairing. Mol. Cell 2007, 27, 91–105.
- Saetrom, P.; Heale, B.S.; Snøve, O., Jr.; Aagaard, L.; Alluin, J.; Rossi, J.J. Distance constraints between microRNA target sites dictate efficacy and cooperativity. Nucleic Acids Res. 2007, 35, 2333–2342.
- Desterro, J.; Bak-Gordon, P.; Carmo-Fonseca, M. Targeting mRNA processing as an anticancer strategy. Nat. Rev. Drug Discov. 2020, 19, 112–129.
- Vogelstein, B.; Papadopoulos, N.; Velculescu, V.E.; Zhou, S.; Diaz, L.A., Jr.; Kinzler, K.W. Cancer genome landscapes. Science 2013, 339, 1546–1558.
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323.
- Ding, J.; Cao, Y.; Qi, C.; Zong, Z. Dysregulated microRNAs participate in the crosstalk between colorectal cancer and atrial fibrillation. Hum. Cell 2023, 36, 1336–1342.
- Wang, H.; Peng, R.; Wang, J.; Qin, Z.; Xue, L. Circulating microRNAs as potential cancer biomarkers: The advantage and disadvantage. Clin. Epigenet. 2018, 10, 59.
- Zhang, L.; Zhang, Y.; Zhao, Y.; Wang, Y.; Ding, H.; Xue, S.; Li, P. Circulating miRNAs as biomarkers for early diagnosis of coronary artery disease. Expert. Opin. Ther. Pat. 2018, 28, 591–601.
- Landgraf, P.; Rusu, M.; Sheridan, R.; Sewer, A.; Iovino, N.; Aravin, A.; Pfeffer, S.; Rice, A.; Kamphorst, A.O.; Landthaler, M.; et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell 2007, 129, 1401–1414.
- Ludwig, N.; Leidinger, P.; Becker, K.; Backes, C.; Fehlmann, T.; Pallasch, C.; Rheinheimer, S.; Meder, B.; Stahler, C.; Meese, E.; et al. Distribution of miRNA expression across human tissues. Nucleic Acids Res. 2016, 44, 3865–3877.
- Reda El Sayed, S.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA Therapeutics in Cancer: Current Advances and Challenges. Cancers 2021, 13, 2680.
- Zhang, S.; Cheng, Z.; Wang, Y.; Han, T. The Risks of miRNA Therapeutics: In a Drug Target Perspective. Drug Des. Devel Ther. 2021, 15, 721–733.
- Mullard, A. 2018 FDA drug approvals. Nat. Rev. Drug Discov. 2019, 18, 85–89.
- Mullard, A. 2019 FDA drug approvals. Nat. Rev. Drug Discov. 2020, 19, 79–84.
- Urits, I.; Swanson, D.; Swett, M.C.; Patel, A.; Berardino, K.; Amgalan, A.; Berger, A.A.; Kassem, H.; Kaye, A.D.; Viswanath, O. A Review of Patisiran (ONPATTRO®) for the Treatment of Polyneuropathy in People with Hereditary Transthyretin Amyloidosis. Neurol. Ther. 2020, 9, 301–315.
- Conceição, I.; González-Duarte, A.; Obici, L.; Schmidt, H.H.; Simoneau, D.; Ong, M.L.; Amass, L. “Red-flag” symptom clusters in transthyretin familial amyloid polyneuropathy. J. Peripher. Nerv. Syst. 2016, 21, 5–9.
- Vormehr, M.; Türeci, Ö.; Sahin, U. Harnessing Tumor Mutations for Truly Individualized Cancer Vaccines. Annu. Rev. Med. 2019, 70, 395–407.
- Rojas, L.A.; Sethna, Z.; Soares, K.C.; Olcese, C.; Pang, N.; Patterson, E.; Lihm, J.; Ceglia, N.; Guasp, P.; Chu, A.; et al. Personalized RNA neoantigen vaccines stimulate T cells in pancreatic cancer. Nature 2023, 618, 144–150.
- Sahin, U.; Derhovanessian, E.; Miller, M.; Kloke, B.P.; Simon, P.; Löwer, M.; Bukur, V.; Tadmor, A.D.; Luxemburger, U.; Schrörs, B.; et al. Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017, 547, 222–226.
- Heil, F.; Hemmi, H.; Hochrein, H.; Ampenberger, F.; Kirschning, C.; Akira, S.; Lipford, G.; Wagner, H.; Bauer, S. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science 2004, 303, 1526–1529.
- Diebold, S.S.; Kaisho, T.; Hemmi, H.; Akira, S.; Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 2004, 303, 1529–1531.
- Lund, O.; Nielsen, M.; Kesmir, C.; Petersen, A.G.; Lundegaard, C.; Worning, P.; Sylvester-Hvid, C.; Lamberth, K.; Røder, G.; Justesen, S.; et al. Definition of supertypes for HLA molecules using clustering of specificity matrices. Immunogenetics 2004, 55, 797–810.
- Kranz, L.M.; Diken, M.; Haas, H.; Kreiter, S.; Loquai, C.; Reuter, K.C.; Meng, M.; Fritz, D.; Vascotto, F.; Hefesha, H.; et al. Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 2016, 534, 396–401.
- Polack, F.P.; Thomas, S.J.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Perez, J.L.; Pérez Marc, G.; Moreira, E.D.; Zerbini, C.; et al. Safety and Efficacy of the BNT162b2 mRNA COVID-19 Vaccine. N. Engl. J. Med. 2020, 383, 2603–2615.
- Cullis, P.R.; Hope, M.J. Lipid Nanoparticle Systems for Enabling Gene Therapies. Mol. Ther. 2017, 25, 1467–1475.
- Chen, Y.; Gao, D.Y.; Huang, L. In vivo delivery of miRNAs for cancer therapy: Challenges and strategies. Adv. Drug Deliv. Rev. 2015, 81, 128–141.
- Antonarakis, E.S.; Lu, C.; Wang, H.; Luber, B.; Nakazawa, M.; Roeser, J.C.; Chen, Y.; Mohammad, T.A.; Chen, Y.; Fedor, H.L.; et al. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N. Engl. J. Med. 2014, 371, 1028–1038.
- Fu, Y.; Chen, J.; Huang, Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 2019, 1, 24.
- Beck, J.D.; Reidenbach, D.; Salomon, N.; Sahin, U.; Türeci, Ö.; Vormehr, M.; Kranz, L.M. mRNA therapeutics in cancer immunotherapy. Mol. Cancer 2021, 20, 69.
- Jivrajani, M.; Nivsarkar, M. Ligand-targeted bacterial minicells: Futuristic nano-sized drug delivery system for the efficient and cost effective delivery of shRNA to cancer cells. Nanomedicine 2016, 12, 2485–2498.
- Iacomino, G. miRNAs: The Road from Bench to Bedside. Genes 2023, 14, 314.
- Dasgupta, I.; Chatterjee, A. Recent Advances in miRNA Delivery Systems. Methods Protoc. 2021, 4, 10.
- Roberts, T.C.; Langer, R.; Wood, M.J.A. Advances in oligonucleotide drug delivery. Nat. Rev. Drug Discov. 2020, 19, 673–694.
- Dosta, P.; Cryer, A.M.; Dion, M.Z.; Shiraishi, T.; Langston, S.P.; Lok, D.; Wang, J.; Harrison, S.; Hatten, T.; Ganno, M.L.; et al. Investigation of the enhanced antitumour potency of STING agonist after conjugation to polymer nanoparticles. Nat. Nanotechnol. 2023, 18, 1351–1363.
- Schultz, B.R.; Chamberlain, J.S. Recombinant adeno-associated virus transduction and integration. Mol. Ther. 2008, 16, 1189–1199.
- Herrera-Carrillo, E.; Liu, Y.P.; Berkhout, B. Improving miRNA Delivery by Optimizing miRNA Expression Cassettes in Diverse Virus Vectors. Hum. Gene Ther. Methods 2017, 28, 177–190.
- Mascellino, M.T.; Di Timoteo, F.; De Angelis, M.; Oliva, A. Overview of the Main Anti-SARS-CoV-2 Vaccines: Mechanism of Action, Efficacy and Safety. Infect. Drug Resist. 2021, 14, 3459–3476.
- Keskin, S.; Brouwers, C.C.; Sogorb-Gonzalez, M.; Martier, R.; Depla, J.A.; Vallès, A.; van Deventer, S.J.; Konstantinova, P.; Evers, M.M. AAV5-miHTT Lowers Huntingtin mRNA and Protein without Off-Target Effects in Patient-Derived Neuronal Cultures and Astrocytes. Mol. Ther. Methods Clin. Dev. 2019, 15, 275–284.
- Miniarikova, J.; Zanella, I.; Huseinovic, A.; van der Zon, T.; Hanemaaijer, E.; Martier, R.; Koornneef, A.; Southwell, A.L.; Hayden, M.R.; van Deventer, S.J.; et al. Design, Characterization, and Lead Selection of Therapeutic miRNAs Targeting Huntingtin for Development of Gene Therapy for Huntington’s Disease. Mol. Ther. Nucleic Acids 2016, 5, e297.
- Samaranch, L.; Blits, B.; San Sebastian, W.; Hadaczek, P.; Bringas, J.; Sudhakar, V.; Macayan, M.; Pivirotto, P.J.; Petry, H.; Bankiewicz, K.S. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. Gene Ther. 2017, 24, 253–261.
- Monahan, P.E.; Négrier, C.; Tarantino, M.; Valentino, L.A.; Mingozzi, F. Emerging Immunogenicity and Genotoxicity Considerations of Adeno-Associated Virus Vector Gene Therapy for Hemophilia. J. Clin. Med. 2021, 10, 2471.
- Blanco, E.; Shen, H.; Ferrari, M. Principles of nanoparticle design for overcoming biological barriers to drug delivery. Nat. Biotechnol. 2015, 33, 941–951.
- Reid, G.; Kao, S.C.; Pavlakis, N.; Brahmbhatt, H.; MacDiarmid, J.; Clarke, S.; Boyer, M.; van Zandwijk, N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics 2016, 8, 1079–1085.
- van Zandwijk, N.; Pavlakis, N.; Kao, S.C.; Linton, A.; Boyer, M.J.; Clarke, S.; Huynh, Y.; Chrzanowska, A.; Fulham, M.J.; Bailey, D.L.; et al. Safety and activity of microRNA-loaded minicells in patients with recurrent malignant pleural mesothelioma: A first-in-man, phase 1, open-label, dose-escalation study. Lancet Oncol. 2017, 18, 1386–1396.
- Elsharkasy, O.M.; Nordin, J.Z.; Hagey, D.W.; de Jong, O.G.; Schiffelers, R.M.; Andaloussi, S.E.; Vader, P. Extracellular vesicles as drug delivery systems: Why and how? Adv. Drug Deliv. Rev. 2020, 159, 332–343.
- Baldari, S.; Di Rocco, G.; Magenta, A.; Picozza, M.; Toietta, G. Extracellular Vesicles-Encapsulated MicroRNA-125b Produced in Genetically Modified Mesenchymal Stromal Cells Inhibits Hepatocellular Carcinoma Cell Proliferation. Cells 2019, 8, 1560.
- Gallant-Behm, C.L.; Piper, J.; Lynch, J.M.; Seto, A.G.; Hong, S.J.; Mustoe, T.A.; Maari, C.; Pestano, L.A.; Dalby, C.M.; Jackson, A.L.; et al. A MicroRNA-29 Mimic (Remlarsen) Represses Extracellular Matrix Expression and Fibroplasia in the Skin. J. Invest. Dermatol. 2019, 139, 1073–1081.
- Zhou, J.; Rossi, J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017, 16, 181–202.
- Russo, V.; Paciocco, A.; Affinito, A.; Roscigno, G.; Fiore, D.; Palma, F.; Galasso, M.; Volinia, S.; Fiorelli, A.; Esposito, C.L.; et al. Aptamer-miR-34c Conjugate Affects Cell Proliferation of Non-Small-Cell Lung Cancer Cells. Mol. Ther. Nucleic Acids 2018, 13, 334–346.
- Huang, Y. Preclinical and Clinical Advances of GalNAc-Decorated Nucleic Acid Therapeutics. Mol. Ther. Nucleic Acids 2017, 6, 116–132.
- Durso, M.; Gaglione, M.; Piras, L.; Mercurio, M.E.; Terreri, S.; Olivieri, M.; Marinelli, L.; Novellino, E.; Incoronato, M.; Grieco, P.; et al. Chemical modifications in the seed region of miRNAs 221/222 increase the silencing performances in gastrointestinal stromal tumor cells. Eur. J. Med. Chem. 2016, 111, 15–25.
- Prakash, T.P.; Graham, M.J.; Yu, J.; Carty, R.; Low, A.; Chappell, A.; Schmidt, K.; Zhao, C.; Aghajan, M.; Murray, H.F.; et al. Targeted delivery of antisense oligonucleotides to hepatocytes using triantennary N-acetyl galactosamine improves potency 10-fold in mice. Nucleic Acids Res. 2014, 42, 8796–8807.
- Biessen, E.A.; Vietsch, H.; Rump, E.T.; Fluiter, K.; Kuiper, J.; Bijsterbosch, M.K.; van Berkel, T.J. Targeted delivery of oligodeoxynucleotides to parenchymal liver cells in vivo. Biochem. J. 1999, 340 Pt. 3, 783–792.
- Drenth, J.P.H.; Schattenberg, J.M. The nonalcoholic steatohepatitis (NASH) drug development graveyard: Established hurdles and planning for future success. Expert. Opin. Investig. Drugs 2020, 29, 1365–1375.
- Kelnar, K.; Peltier, H.J.; Leatherbury, N.; Stoudemire, J.; Bader, A.G. Quantification of therapeutic miRNA mimics in whole blood from nonhuman primates. Anal. Chem. 2014, 86, 1534–1542.
- van der Ree, M.H.; de Vree, J.M.; Stelma, F.; Willemse, S.; van der Valk, M.; Rietdijk, S.; Molenkamp, R.; Schinkel, J.; van Nuenen, A.C.; Beuers, U.; et al. Safety, tolerability, and antiviral effect of RG-101 in patients with chronic hepatitis C: A phase 1B, double-blind, randomised controlled trial. Lancet 2017, 389, 709–717.
- Stelma, F.; van der Ree, M.H.; Sinnige, M.J.; Brown, A.; Swadling, L.; de Vree, J.M.L.; Willemse, S.B.; van der Valk, M.; Grint, P.; Neben, S.; et al. Immune phenotype and function of natural killer and T cells in chronic hepatitis C patients who received a single dose of anti-MicroRNA-122, RG-101. Hepatology 2017, 66, 57–68.
- Yamamoto, T.; Mukai, Y.; Wada, F.; Terada, C.; Kayaba, Y.; Oh, K.; Yamayoshi, A.; Obika, S.; Harada-Shiba, M. Highly Potent GalNAc-Conjugated Tiny LNA Anti-miRNA-122 Antisense Oligonucleotides. Pharmaceutics 2021, 13, 817.
- Kardani, A.; Yaghoobi, H.; Alibakhshi, A.; Khatami, M. Inhibition of miR-155 in MCF-7 breast cancer cell line by gold nanoparticles functionalized with antagomir and AS1411 aptamer. J. Cell. Physiol. 2020, 235, 6887–6895.
- Nikolova, M.P.; Chavali, M.S. Recent advances in biomaterials for 3D scaffolds: A review. Bioact. Mater. 2019, 4, 271–292.
- Aguilar-de-Leyva, Á.; Linares, V.; Casas, M.; Caraballo, I. 3D Printed Drug Delivery Systems Based on Natural Products. Pharmaceutics 2020, 12, 620.
- Shende, P.; Trivedi, R. 3D Printed Bioconstructs: Regenerative Modulation for Genetic Expression. Stem Cell Rev. Rep. 2021, 17, 1239–1250.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Entry Collection:
Biopharmaceuticals Technology
Revisions:
2 times
(View History)
Update Date:
30 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




