
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Bi-Dar Wang | -- | 4383 | 2024-01-25 23:45:41 | | | |
| 2 | Mona Zou | -3 word(s) | 4380 | 2024-01-26 10:27:53 | | |
Video Upload Options
Prostate cancer (PCa) has been known as the most prevalent cancer disease and the second leading cause of cancer mortality in men almost all over the globe. There is an urgent need for establishment of PCa models that can recapitulate the progress of genomic landscapes and molecular alterations during development and progression of this disease. Notably, several organoid models have been developed for assessing the complex interaction between PCa and its surrounding microenvironment. PCa organoids have been emerged as powerful in vitro 3D model systems that recapitulate the molecular features (such as genomic/epigenomic changes and tumor microenvironment) of PCa metastatic tumors. In addition, application of organoid technology in mechanistic studies (i.e., for understanding cellular/subcellular and molecular alterations) and translational medicine has been recognized as a promising approach for facilitating the development of potential biomarkers and novel therapeutic strategies.
1. Background and Recent Stage of Prostate Organoid Culture
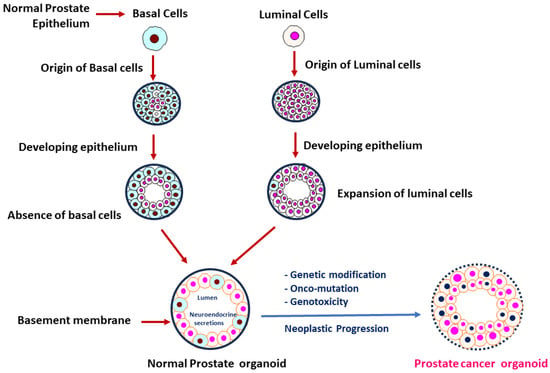
2. Tumor Model System in PCa
2.1. In Vitro and In Vivo 3D PCa Models
2.1.1. Spheroids
2.1.2. Patient-Derived Organoids (PDOs)
2.1.3. Induced-Pluripotent Stem Cell (iPSC)-Derived Organoids
2.1.4. Patient-Derived Xenograft (PDX)-Derived Organoids
2.1.5. Three-Dimensional Models for PCa Epigenomic Studies
2.1.6. Strategies for PDO Development from Tumor Immune Microenvironment (TIME)
2.1.7. In Vivo Implantation of PCa Organoids
References
- Shi, X.; Gipp, J.; Bushman, W. Anchorage-independent culture maintains prostate stem cells. Dev. Biol. 2007, 312, 396–406.
- Drost, J.; Karthaus, W.R.; Gao, D.; Driehuis, E.; Sawyers, C.L.; Chen, Y.; Clevers, H. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 2016, 11, 347–358.
- Lukacs, R.U.; Goldstein, A.S.; Lawson, D.A.; Cheng, D.; Witte, O.N. Isolation, cultivation and characterization of adult murine prostate stem cells. Nat. Protoc. 2010, 5, 702–713.
- Karthaus, W.R.; Iaquinta, P.J.; Drost, J.; Gracanin, A.; van Boxtel, R.; Wongvipat, J.; Dowling, C.M.; Gao, D.; Begthel, H.; Sachs, N.; et al. Identification of multipotent luminal progenitor cells in human prostate organoid cultures. Cell 2014, 159, 163–175.
- Karkampouna, S.; La Manna, F.; Benjak, A.; Kiener, M.; De Menna, M.; Zoni, E.; Grosjean, J.; Klima, I.; Garofoli, A.; Bolis, M.; et al. Patient-derived xenografts and organoids model therapy response in prostate cancer. Nat. Commun. 2021, 12, 1117.
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265.
- Jung, P.; Sato, T.; Merlos-Suarez, A.; Barriga, F.M.; Iglesias, M.; Rossell, D.; Auer, H.; Gallardo, M.; Blasco, M.A.; Sancho, E.; et al. Isolation and in vitro expansion of human colonic stem cells. Nat. Med. 2011, 17, 1225–1227.
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid cultures derived from patients with advanced prostate cancer. Cell 2014, 159, 176–187.
- Drost, J.; Clevers, H. Organoids in cancer research. Nat. Rev. Cancer 2018, 18, 407–418.
- Beshiri, M.; Agarwal, S.; Yin, J.J.; Kelly, K. Prostate organoids: Emerging experimental tools for translational research. J. Clin. Investig. 2023, 133, e169616.
- Chua, C.W.; Shibata, M.; Lei, M.; Toivanen, R.; Barlow, L.J.; Bergren, S.K.; Badani, K.K.; McKiernan, J.M.; Benson, M.C.; Hibshoosh, H.; et al. Single luminal epithelial progenitors can generate prostate organoids in culture. Nat. Cell Biol. 2014, 16, 951–961.
- Agarwal, S.; Hynes, P.G.; Tillman, H.S.; Lake, R.; Abou-Kheir, W.G.; Fang, L.; Casey, O.M.; Ameri, A.H.; Martin, P.L.; Yin, J.J.; et al. Identification of Different Classes of Luminal Progenitor Cells within Prostate Tumors. Cell Rep. 2015, 13, 2147–2158.
- Beshiri, M.L.; Tice, C.M.; Tran, C.; Nguyen, H.M.; Sowalsky, A.G.; Agarwal, S.; Jansson, K.H.; Yang, Q.; McGowen, K.M.; Yin, J.; et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin. Cancer Res. 2018, 24, 4332–4345.
- Puca, L.; Bareja, R.; Prandi, D.; Shaw, R.; Benelli, M.; Karthaus, W.R.; Hess, J.; Sigouros, M.; Donoghue, A.; Kossai, M.; et al. Patient derived organoids to model rare prostate cancer phenotypes. Nat. Commun. 2018, 9, 2404.
- Tang, F.; Xu, D.; Wang, S.; Wong, C.K.; Martinez-Fundichely, A.; Lee, C.J.; Cohen, S.; Park, J.; Hill, C.E.; Eng, K.; et al. Chromatin profiles classify castration-resistant prostate cancers suggesting therapeutic targets. Science 2022, 376, eabe1505.
- Sartor, O.; de Bono, J.S. Metastatic Prostate Cancer. N. Engl. J. Med. 2018, 378, 645–657.
- Risbridger, G.P.; Toivanen, R.; Taylor, R.A. Preclinical Models of Prostate Cancer: Patient-Derived Xenografts, Organoids, and Other Explant Models. Cold Spring Harb. Perspect. Med. 2018, 8, a030536.
- Beltran, H.; Oromendia, C.; Danila, D.C.; Montgomery, B.; Hoimes, C.; Szmulewitz, R.Z.; Vaishampayan, U.; Armstrong, A.J.; Stein, M.; Pinski, J.; et al. A Phase II Trial of the Aurora Kinase A Inhibitor Alisertib for Patients with Castration-resistant and Neuroendocrine Prostate Cancer: Efficacy and Biomarkers. Clin. Cancer Res. 2019, 25, 43–51.
- Watt, M.J.; Clark, A.K.; Selth, L.A.; Haynes, V.R.; Lister, N.; Rebello, R.; Porter, L.H.; Niranjan, B.; Whitby, S.T.; Lo, J.; et al. Suppressing fatty acid uptake has therapeutic effects in preclinical models of prostate cancer. Sci. Transl. Med. 2019, 11, eaau5758.
- Chakraborty, G.; Armenia, J.; Mazzu, Y.Z.; Nandakumar, S.; Stopsack, K.H.; Atiq, M.O.; Komura, K.; Jehane, L.; Hirani, R.; Chadalavada, K.; et al. Significance of BRCA2 and RB1 Co-loss in Aggressive Prostate Cancer Progression. Clin. Cancer Res. 2020, 26, 2047–2064.
- Hwang, J.H.; Seo, J.H.; Beshiri, M.L.; Wankowicz, S.; Liu, D.; Cheung, A.; Li, J.; Qiu, X.; Hong, A.L.; Botta, G.; et al. CREB5 Promotes Resistance to Androgen-Receptor Antagonists and Androgen Deprivation in Prostate Cancer. Cell Rep. 2019, 29, 2355–2370.e6.
- Zhang, Z.; Karthaus, W.R.; Lee, Y.S.; Gao, V.R.; Wu, C.; Russo, J.W.; Liu, M.; Mota, J.M.; Abida, W.; Linton, E.; et al. Tumor Microenvironment-Derived NRG1 Promotes Antiandrogen Resistance in Prostate Cancer. Cancer Cell 2020, 38, 279–296.e9.
- Joshi, A.; Roberts, M.J.; Alinezhad, S.; Williams, E.D.; Vela, I. Challenges, applications and future directions of precision medicine in prostate cancer—The role of organoids and patient-derived xenografts. BJU Int. 2020, 126, 65–72.
- Gil, V.; Miranda, S.; Riisnaes, R.; Gurel, B.; D’Ambrosio, M.; Vasciaveo, A.; Crespo, M.; Ferreira, A.; Brina, D.; Troiani, M.; et al. HER3 Is an Actionable Target in Advanced Prostate Cancer. Cancer Res. 2021, 81, 6207–6218.
- Heninger, E.; Kosoff, D.; Rodems, T.S.; Sethakorn, N.; Singh, A.; Gungurthi, H.; Carlson, K.N.; Yang, B.; Gilsdorf, C.; Pasch, C.A.; et al. Live cell molecular analysis of primary prostate cancer organoids identifies persistent androgen receptor signaling. Med. Oncol. 2021, 38, 135.
- Grbesa, I.; Augello, M.A.; Liu, D.; McNally, D.R.; Gaffney, C.D.; Huang, D.; Lin, K.; Ivenitsky, D.; Goueli, R.; Robinson, B.D.; et al. Reshaping of the androgen-driven chromatin landscape in normal prostate cells by early cancer drivers and effect on therapeutic sensitivity. Cell Rep. 2021, 36, 109625.
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597.
- Pamarthy, S.; Sabaawy, H.E. Patient derived organoids in prostate cancer: Improving therapeutic efficacy in precision medicine. Mol. Cancer 2021, 20, 125.
- Weiswald, L.B.; Bellet, D.; Dangles-Marie, V. Spherical cancer models in tumor biology. Neoplasia 2015, 17, 1–15.
- Martinez, S.R.; Elix, C.C.; Ochoa, P.T.; Sanchez-Hernandez, E.S.; Alkashgari, H.R.; Ortiz-Hernandez, G.L.; Zhang, L.; Casiano, C.A. Glucocorticoid Receptor and beta-Catenin Interact in Prostate Cancer Cells and Their Co-Inhibition Attenuates Tumorsphere Formation, Stemness, and Docetaxel Resistance. Int. J. Mol. Sci. 2023, 24, 7130.
- Thulin, M.H.; Maatta, J.; Linder, A.; Sterbova, S.; Ohlsson, C.; Damber, J.E.; Widmark, A.; Persson, E. Inhibition of STAT3 prevents bone metastatic progression of prostate cancer in vivo. Prostate 2021, 81, 452–462.
- Bialkowska, K.; Komorowski, P.; Bryszewska, M.; Milowska, K. Spheroids as a Type of Three-Dimensional Cell Cultures-Examples of Methods of Preparation and the Most Important Application. Int. J. Mol. Sci. 2020, 21, 6225.
- Ellem, S.J.; De-Juan-Pardo, E.M.; Risbridger, G.P. In vitro modeling of the prostate cancer microenvironment. Adv. Drug Deliv. Rev. 2014, 79–80, 214–221.
- Payne, M.C.; Ho, S.; Hashimoto, T.; Imboden, S.; Diaz, J.A.; Lee, B.S.; Rupert, M.J.; Cai, N.Y.; Goldstein, A.S.; Lin, N.Y.C. Microwell-based flow culture increases viability and restores drug response in prostate cancer spheroids. Biotechnol. J. 2023, 18, e2200434.
- Sato, T.; Stange, D.E.; Ferrante, M.; Vries, R.G.; Van Es, J.H.; Van den Brink, S.; Van Houdt, W.J.; Pronk, A.; Van Gorp, J.; Siersema, P.D.; et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology 2011, 141, 1762–1772.
- Kashfi, S.M.H.; Almozyan, S.; Jinks, N.; Koo, B.K.; Nateri, A.S. Morphological alterations of cultured human colorectal matched tumour and healthy organoids. Oncotarget 2018, 9, 10572–10584.
- Behjati, S.; Huch, M.; van Boxtel, R.; Karthaus, W.; Wedge, D.C.; Tamuri, A.U.; Martincorena, I.; Petljak, M.; Alexandrov, L.B.; Gundem, G.; et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 2014, 513, 422–425.
- Boj, S.F.; Hwang, C.I.; Baker, L.A.; Chio, I.I.; Engle, D.D.; Corbo, V.; Jager, M.; Ponz-Sarvise, M.; Tiriac, H.; Spector, M.S.; et al. Organoid models of human and mouse ductal pancreatic cancer. Cell 2015, 160, 324–338.
- Ishiguro, T.; Sato, A.; Ohata, H.; Ikarashi, Y.; Takahashi, R.U.; Ochiya, T.; Yoshida, M.; Tsuda, H.; Onda, T.; Kato, T.; et al. Establishment and Characterization of an In Vitro Model of Ovarian Cancer Stem-like Cells with an Enhanced Proliferative Capacity. Cancer Res. 2016, 76, 150–160.
- Ohata, H.; Ishiguro, T.; Aihara, Y.; Sato, A.; Sakai, H.; Sekine, S.; Taniguchi, H.; Akasu, T.; Fujita, S.; Nakagama, H.; et al. Induction of the stem-like cell regulator CD44 by Rho kinase inhibition contributes to the maintenance of colon cancer-initiating cells. Cancer Res. 2012, 72, 5101–5110.
- Linxweiler, J.; Hammer, M.; Muhs, S.; Kohn, M.; Pryalukhin, A.; Veith, C.; Bohle, R.M.; Stockle, M.; Junker, K.; Saar, M. Patient-derived, three-dimensional spheroid cultures provide a versatile translational model for the study of organ-confined prostate cancer. J. Cancer Res. Clin. Oncol. 2019, 145, 551–559.
- Moad, M.; Pal, D.; Hepburn, A.C.; Williamson, S.C.; Wilson, L.; Lako, M.; Armstrong, L.; Hayward, S.W.; Franco, O.E.; Cates, J.M.; et al. A novel model of urinary tract differentiation, tissue regeneration, and disease: Reprogramming human prostate and bladder cells into induced pluripotent stem cells. Eur. Urol. 2013, 64, 753–761.
- Hepburn, A.C.; Curry, E.L.; Moad, M.; Steele, R.E.; Franco, O.E.; Wilson, L.; Singh, P.; Buskin, A.; Crawford, S.E.; Gaughan, L.; et al. Propagation of human prostate tissue from induced pluripotent stem cells. Stem Cells Transl. Med. 2020, 9, 734–745.
- Hepburn, A.C.; Curry, E.L.; Moad, M.; Steele, R.E.; Franco, O.E.; Wilson, L.; Singh, P.; Crawford, S.; Gaughan, L.; Mills, I.; et al. High-throughput propagation of human prostate tissue from induced-pluripotent stem cells. bioRxiv 2019, bioRxiv:637876.
- Breunig, M.; Merkle, J.; Wagner, M.; Melzer, M.K.; Barth, T.F.E.; Engleitner, T.; Krumm, J.; Wiedenmann, S.; Cohrs, C.M.; Perkhofer, L.; et al. Modeling plasticity and dysplasia of pancreatic ductal organoids derived from human pluripotent stem cells. Cell Stem Cell 2021, 28, 1105–1124.e19.
- Sancho-Martinez, I.; Nivet, E.; Xia, Y.; Hishida, T.; Aguirre, A.; Ocampo, A.; Ma, L.; Morey, R.; Krause, M.N.; Zembrzycki, A.; et al. Establishment of human iPSC-based models for the study and targeting of glioma initiating cells. Nat. Commun. 2016, 7, 10743.
- Feng, W.; Cao, Z.; Lim, P.X.; Zhao, H.; Luo, H.; Mao, N.; Lee, Y.S.; Rivera, A.A.; Choi, D.; Wu, C.; et al. Rapid interrogation of cancer cell of origin through CRISPR editing. Proc. Natl. Acad. Sci. USA 2021, 118, e2110344118.
- Schutgens, F.; Clevers, H. Human Organoids: Tools for Understanding Biology and Treating Diseases. Annu. Rev. Pathol. 2020, 15, 211–234.
- Shi, C.; Chen, X.; Tan, D. Development of patient-derived xenograft models of prostate cancer for maintaining tumor heterogeneity. Transl. Androl. Urol. 2019, 8, 519–528.
- Basak, D.; Gregori, L.; Johora, F.; Deb, S. Preclinical and Clinical Research Models of Prostate Cancer: A Brief Overview. Life 2022, 12, 1607.
- Aparicio, S.; Hidalgo, M.; Kung, A.L. Examining the utility of patient-derived xenograft mouse models. Nat. Rev. Cancer 2015, 15, 311–316.
- Stevens, C.; Hightower, A.; Buxbaum, S.G.; Falzarano, S.M.; Rhie, S.K. Genomic, epigenomic, and transcriptomic signatures of prostate cancer between African American and European American patients. Front. Oncol. 2023, 13, 1079037.
- Conteduca, V.; Hess, J.; Yamada, Y.; Ku, S.Y.; Beltran, H. Epigenetics in prostate cancer: Clinical implications. Transl. Androl. Urol. 2021, 10, 3104–3116.
- Saghafinia, S.; Mina, M.; Riggi, N.; Hanahan, D.; Ciriello, G. Pan-Cancer Landscape of Aberrant DNA Methylation across Human Tumors. Cell Rep. 2018, 25, 1066–1080.e8.
- Cancer Genome Atlas Research, N. The Molecular Taxonomy of Primary Prostate Cancer. Cell 2015, 163, 1011–1025.
- Zavridou, M.; Strati, A.; Bournakis, E.; Smilkou, S.; Tserpeli, V.; Lianidou, E. Prognostic Significance of Gene Expression and DNA Methylation Markers in Circulating Tumor Cells and Paired Plasma Derived Exosomes in Metastatic Castration Resistant Prostate Cancer. Cancers 2021, 13, 780.
- Zhao, S.G.; Chen, W.S.; Li, H.; Foye, A.; Zhang, M.; Sjostrom, M.; Aggarwal, R.; Playdle, D.; Liao, A.; Alumkal, J.J.; et al. The DNA methylation landscape of advanced prostate cancer. Nat. Genet. 2020, 52, 778–789.
- Lu, A.; Wang, J.; Sun, W.; Huang, W.; Cai, Z.; Zhao, G.; Wang, J. Reprogrammable CRISPR/dCas9-based recruitment of DNMT1 for site-specific DNA demethylation and gene regulation. Cell Discov. 2019, 5, 22.
- Stepper, P.; Kungulovski, G.; Jurkowska, R.Z.; Chandra, T.; Krueger, F.; Reinhardt, R.; Reik, W.; Jeltsch, A.; Jurkowski, T.P. Efficient targeted DNA methylation with chimeric dCas9-Dnmt3a-Dnmt3L methyltransferase. Nucleic Acids Res. 2017, 45, 1703–1713.
- Pomerantz, M.M.; Qiu, X.; Zhu, Y.; Takeda, D.Y.; Pan, W.; Baca, S.C.; Gusev, A.; Korthauer, K.D.; Severson, T.M.; Ha, G.; et al. Prostate cancer reactivates developmental epigenomic programs during metastatic progression. Nat. Genet. 2020, 52, 790–799.
- Duan, L.; Chen, Z.; Lu, J.; Liang, Y.; Wang, M.; Roggero, C.M.; Zhang, Q.J.; Gao, J.; Fang, Y.; Cao, J.; et al. Histone lysine demethylase KDM4B regulates the alternative splicing of the androgen receptor in response to androgen deprivation. Nucleic Acids Res. 2019, 47, 11623–11636.
- Wang, N.; Ma, T.; Yu, B. Targeting epigenetic regulators to overcome drug resistance in cancers. Signal Transduct. Target. Ther. 2023, 8, 69.
- Hilton, I.B.; D’Ippolito, A.M.; Vockley, C.M.; Thakore, P.I.; Crawford, G.E.; Reddy, T.E.; Gersbach, C.A. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nat. Biotechnol. 2015, 33, 510–517.
- Elbadawy, M.; Abugomaa, A.; Yamawaki, H.; Usui, T.; Sasaki, K. Development of Prostate Cancer Organoid Culture Models in Basic Medicine and Translational Research. Cancers 2020, 12, 777.
- Driehuis, E.; Clevers, H. CRISPR/Cas 9 genome editing and its applications in organoids. Am. J. Physiol. Gastrointest. Liver Physiol. 2017, 312, G257–G265.
- Davidsson, S.; Ohlson, A.L.; Andersson, S.O.; Fall, K.; Meisner, A.; Fiorentino, M.; Andren, O.; Rider, J.R. CD4 helper T cells, CD8 cytotoxic T cells, and FOXP3+ regulatory T cells with respect to lethal prostate cancer. Mod. Pathol. 2013, 26, 448–455.
- Vitkin, N.; Nersesian, S.; Siemens, D.R.; Koti, M. The Tumor Immune Contexture of Prostate Cancer. Front. Immunol. 2019, 10, 603.
- Movassaghi, M.; Chung, R.; Anderson, C.B.; Stein, M.; Saenger, Y.; Faiena, I. Overcoming Immune Resistance in Prostate Cancer: Challenges and Advances. Cancers 2021, 13, 4757.
- Tuong, Z.K.; Loudon, K.W.; Berry, B.; Richoz, N.; Jones, J.; Tan, X.; Nguyen, Q.; George, A.; Hori, S.; Field, S.; et al. Resolving the immune landscape of human prostate at a single-cell level in health and cancer. Cell Rep. 2021, 37, 110132.
- Guan, X.; Polesso, F.; Wang, C.; Sehrawat, A.; Hawkins, R.M.; Murray, S.E.; Thomas, G.V.; Caruso, B.; Thompson, R.F.; Wood, M.A.; et al. Androgen receptor activity in T cells limits checkpoint blockade efficacy. Nature 2022, 606, 791–796.
- Nair, S.S.; Weil, R.; Dovey, Z.; Davis, A.; Tewari, A.K. The Tumor Microenvironment and Immunotherapy in Prostate and Bladder Cancer. Urol. Clin. N. Am. 2020, 47, e17–e54.
- Antonarakis, E.S.; Piulats, J.M.; Gross-Goupil, M.; Goh, J.; Ojamaa, K.; Hoimes, C.J.; Vaishampayan, U.; Berger, R.; Sezer, A.; Alanko, T.; et al. Pembrolizumab for Treatment-Refractory Metastatic Castration-Resistant Prostate Cancer: Multicohort, Open-Label Phase II KEYNOTE-199 Study. J. Clin. Oncol. 2020, 38, 395–405.
- Cha, H.R.; Lee, J.H.; Ponnazhagan, S. Revisiting Immunotherapy: A Focus on Prostate Cancer. Cancer Res. 2020, 80, 1615–1623.
- Masopust, D.; Sivula, C.P.; Jameson, S.C. Of Mice, Dirty Mice, and Men: Using Mice To Understand Human Immunology. J. Immunol. 2017, 199, 383–388.
- Sun, L.; Li, C.W.; Chung, E.M.; Yang, R.; Kim, Y.S.; Park, A.H.; Lai, Y.J.; Yang, Y.; Wang, Y.H.; Liu, J.; et al. Targeting Glycosylated PD-1 Induces Potent Antitumor Immunity. Cancer Res. 2020, 80, 2298–2310.
- Lee, J.K.; Bangayan, N.J.; Chai, T.; Smith, B.A.; Pariva, T.E.; Yun, S.; Vashisht, A.; Zhang, Q.; Park, J.W.; Corey, E.; et al. Systemic surfaceome profiling identifies target antigens for immune-based therapy in subtypes of advanced prostate cancer. Proc. Natl. Acad. Sci. USA 2018, 115, E4473–E4482.
- Yamaguchi, Y.; Gibson, J.; Ou, K.; Lopez, L.S.; Ng, R.H.; Leggett, N.; Jonsson, V.D.; Zarif, J.C.; Lee, P.P.; Wang, X.; et al. PD-L1 blockade restores CAR T cell activity through IFN-gamma-regulation of CD163+ M2 macrophages. J. Immunother. Cancer 2022, 10, e004400.
- Rodrigues, J.; Heinrich, M.A.; Teixeira, L.M.; Prakash, J. 3D In Vitro Model (R)evolution: Unveiling Tumor-Stroma Interactions. Trends Cancer 2021, 7, 249–264.
- Colombo, E.; Cattaneo, M.G. Multicellular 3D Models to Study Tumour-Stroma Interactions. Int. J. Mol. Sci. 2021, 22, 1633.
- Jeong, S.R.; Kang, M. Exploring Tumor-Immune Interactions in Co-Culture Models of T Cells and Tumor Organoids Derived from Patients. Int. J. Mol. Sci. 2023, 24, 14609.
- Sun, C.P.; Lan, H.R.; Fang, X.L.; Yang, X.Y.; Jin, K.T. Organoid Models for Precision Cancer Immunotherapy. Front. Immunol. 2022, 13, 770465.
- Yuan, J.; Li, X.; Yu, S. Cancer organoid co-culture model system: Novel approach to guide precision medicine. Front. Immunol. 2022, 13, 1061388.
- Neal, J.T.; Li, X.; Zhu, J.; Giangarra, V.; Grzeskowiak, C.L.; Ju, J.; Liu, I.H.; Chiou, S.H.; Salahudeen, A.A.; Smith, A.R.; et al. Organoid Modeling of the Tumor Immune Microenvironment. Cell 2018, 175, 1972–1988.e16.
- Ou, L.; Liu, S.; Wang, H.; Guo, Y.; Guan, L.; Shen, L.; Luo, R.; Elder, D.E.; Huang, A.C.; Karakousis, G.; et al. Patient-derived melanoma organoid models facilitate the assessment of immunotherapies. EBioMedicine 2023, 92, 104614.
- Tsai, S.; McOlash, L.; Palen, K.; Johnson, B.; Duris, C.; Yang, Q.; Dwinell, M.B.; Hunt, B.; Evans, D.B.; Gershan, J.; et al. Development of primary human pancreatic cancer organoids, matched stromal and immune cells and 3D tumor microenvironment models. BMC Cancer 2018, 18, 335.
- Piro, G.; Agostini, A.; Larghi, A.; Quero, G.; Carbone, C.; Esposito, A.; Rizzatti, G.; Attili, F.; Alfieri, S.; Costamagna, G.; et al. Pancreatic Cancer Patient-Derived Organoid Platforms: A Clinical Tool to Study Cell- and Non-Cell-Autonomous Mechanisms of Treatment Response. Front. Med. 2021, 8, 793144.
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12.
- Fan, T.W.; Higashi, R.M.; Song, H.; Daneshmandi, S.; Mahan, A.L.; Purdom, M.S.; Bocklage, T.J.; Pittman, T.A.; He, D.; Wang, C.; et al. Innate immune activation by checkpoint inhibition in human patient-derived lung cancer tissues. Elife 2021, 10, e69578.
- Zhou, Y.; Lin, Z.; Zhang, X.; Chen, C.; Zhao, H.; Hong, S.; Zhang, L. First-line treatment for patients with advanced non-small cell lung carcinoma and high PD-L1 expression: Pembrolizumab or pembrolizumab plus chemotherapy. J. Immunother. Cancer 2019, 7, 120.
- Tanaka, C.; Furihata, K.; Naganuma, S.; Ogasawara, M.; Yoshioka, R.; Taniguchi, H.; Furihata, M.; Taniuchi, K. Establishment of a mouse model of pancreatic cancer using human pancreatic cancer cell line S2-013-derived organoid. Hum. Cell 2022, 35, 735–744.
- Roper, J.; Tammela, T.; Cetinbas, N.M.; Akkad, A.; Roghanian, A.; Rickelt, S.; Almeqdadi, M.; Wu, K.; Oberli, M.A.; Sanchez-Rivera, F.J.; et al. In vivo genome editing and organoid transplantation models of colorectal cancer and metastasis. Nat. Biotechnol. 2017, 35, 569–576.
- Park, J.W.; Lee, J.K.; Phillips, J.W.; Huang, P.; Cheng, D.; Huang, J.; Witte, O.N. Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc. Natl. Acad. Sci. USA 2016, 113, 4482–4487.
- Shultz, L.D.; Ishikawa, F.; Greiner, D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007, 7, 118–130.
- Cambuli, F.; Foletto, V.; Alaimo, A.; De Felice, D.; Gandolfi, F.; Palumbieri, M.D.; Zaffagni, M.; Genovesi, S.; Lorenzoni, M.; Celotti, M.; et al. Intra-epithelial non-canonical Activin A signaling safeguards prostate progenitor quiescence. EMBO Rep. 2022, 23, e54049.




