You're using an outdated browser. Please upgrade to a modern browser for the best experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jian Zha | -- | 3420 | 2024-01-17 14:49:45 | | | |
| 2 | Lindsay Dong | Meta information modification | 3420 | 2024-01-19 03:08:22 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zha, J.; Liu, D.; Ren, J.; Liu, Z.; Wu, X. Pichia pastoris Strains as Powerful Cell Factories. Encyclopedia. Available online: https://encyclopedia.pub/entry/53976 (accessed on 23 December 2025).
Zha J, Liu D, Ren J, Liu Z, Wu X. Pichia pastoris Strains as Powerful Cell Factories. Encyclopedia. Available at: https://encyclopedia.pub/entry/53976. Accessed December 23, 2025.
Zha, Jian, Dan Liu, Juan Ren, Zhijun Liu, Xia Wu. "Pichia pastoris Strains as Powerful Cell Factories" Encyclopedia, https://encyclopedia.pub/entry/53976 (accessed December 23, 2025).
Zha, J., Liu, D., Ren, J., Liu, Z., & Wu, X. (2024, January 17). Pichia pastoris Strains as Powerful Cell Factories. In Encyclopedia. https://encyclopedia.pub/entry/53976
Zha, Jian, et al. "Pichia pastoris Strains as Powerful Cell Factories." Encyclopedia. Web. 17 January, 2024.
Copy Citation
Pichia pastoris is the most widely used microorganism for the production of secreted industrial proteins and therapeutic proteins. This yeast has been repurposed as a cell factory for the production of chemicals and natural products.
Pichia pastoris
natural product
cell factory
1. Introduction
Methylotrophic yeasts are microorganisms capable of utilizing methanol as the sole source of carbon and energy and are hence considered as potential hosts for green bio-manufacturing. Meanwhile, these microbes, when used as cell factories, have some other advantages such as high fermentation density, low accumulation of toxic metabolites, ability to grow in cheap basal salt media, and capability of complete protein modifications and processing.
Pichia pastoris, which has been renamed as Komagataella phaffii [1], is one of the typical methylotrophic yeasts and plays an important role in the production of recombinant proteins. Using the P. pastoris expression system, thousands of recombinant proteins have been synthesized so far, some of which have successfully entered the market, such as human insulin and human interferon-α [2].
P. pastoris is a unicellular microorganism with the advantages of high expression levels of recombinant proteins, ease of large-scale cultivation, and low cultivation costs [3]. Its cell density can reach 130 g/L during industrial production [4], and the yield of recombinant proteins can reach tens of grams per liter [5]. Recombinant proteins can be expressed both intracellularly and in a secreted form in P. pastoris. The efficiency of secretory expression in P. pastoris is generally higher than that in Saccharomyces cerevisiae, which is another commonly adopted workhorse for microbial bio-production [6]. P. pastoris has a clear genetic background and is excellent in protein folding and post-translational modifications, especially glycosylation [7][8]. In P. pastoris, oligosaccharide chains added to proteins after translation (8–14 mannose residues per side chain) are shorter than in S. cerevisiae (50–150 mannose residues), and O-linked glycosylation is very minimal, potentially avoiding the risk of excessive glycosylation (Figure 1) [9]. Moreover, P. pastoris contains a strong methanol utilization pathway and is able to provide some key cofactors [10]. Another major difference between these two yeasts is that the homologous recombination (HR) pathway in P. pastoris is significantly weaker than the non-homologous end joining (NHEJ) pathway [11].
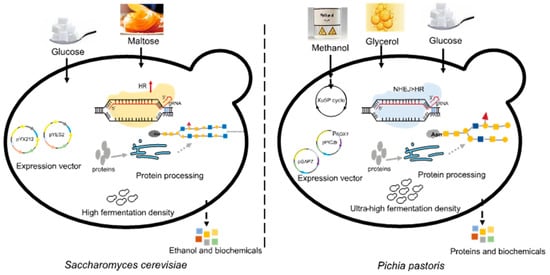
Figure 1. Differences between Pichia pastoris and Saccharomyces cerevisiae.
2. Physiological Characteristics of Pichia pastoris
P. pastoris belongs to the Ascomycetes class [12]. Its colony is generally milk-white with a smooth surface, showing a bulge (Figure 2). This yeast mainly exists in the haploid form in the asexual growth phase. Under nutrient limitation, two haploid cells can be induced with different physiological types to mate and fuse into diploids [13].
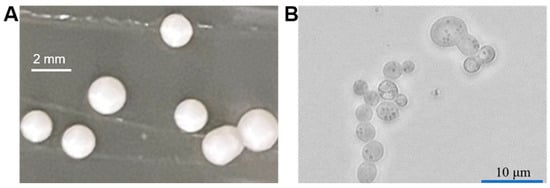
Figure 2. Colony morphology (A) and microscopic image (B) of P. pastoris.
P. pastoris grows optimally at 28–30 °C [14] with a tolerance of pH ranging from 3 to 7 [15]. The carbon source includes a variety of compounds such as glucose [16], glycerol [17], L-rhamnose [18], formate [19], and so on. This microbe can also use methanol as its sole source of carbon and energy owing to the presence of enzymes in the peroxisomes essential to methanol metabolism, such as alcohol oxidase, dihydroxyacetone synthase, and peroxidase [20]. Among these, the expression of alcohol oxidase is strictly induced and controlled by methanol, so the enzyme activity can only be detected in the presence of methanol [21]. P. pastoris can use ammonium sulfate, proline, and peptone as the nitrogen source. When ammonium sulfate is adopted, the expression levels of major genes related to methanol utilization (gene MUT) and peroxisome biogenesis/degradation (gene PEX) are the highest [22]. Meanwhile, an appropriate NH4+ concentration is beneficial for cell growth and heterologous protein expression [22][23]. In some cases, the use of complex nitrogen sources such as casamino acids and peptone can effectively alleviate proteolytic degradation of heterologous proteins [24].
3. The Pichia pastoris Expression System
3.1. Strains
At present, the commonly used P. pastoris strains for heterologous protein expression include X-33, GS115, KM71, SMD1163, and MC100-3. The characteristics of these strains have been described in detail in other reports [25][26]. Based on their ability to utilize methanol, P. pastoris strains are mainly divided into three phenotypes, i.e., Mut+, MutS, and Mut−. Strain X-33 is a wild-type strain carrying genes AOX1 and AOX2 (both encoding alcohol oxidase) and the phenotype is Mut+ (methanol utilization plus), which is often used to express recombinant plasmids containing zeocin resistance. Both GS115 and KM71 strains are histidine deficient. The GS115 strain contains AOX1 and AOX2, and its own phenotype is Mut+. In comparison, the KM71 strain is MutS (methanol utilization slow) because the gene AOX1 is replaced by the S. cerevisiae Arg4, and the strain can only utilize methanol at a slower rate dependent on the weakly controlled gene AOX2. Strain MC100-3 is deficient in both AOX genes and hence cannot grow on methanol, thus performing the Mut− (methanol utilization minus) phenotype. SMD1163 is a protease deficient strain in which the pep4 gene encoding protease A and the prb1 gene encoding a subtilisin-like protease are knocked out. This strain is favorable for the expression of heterologous proteins sensitive to proteases [27].
3.2. Expression Vectors
According to the location of the expressed proteins, P. pastoris vectors can be divided into intracellular expression vectors (such as pPIC3, pPICZ, pPHIL-D2, etc.) and secretory expression vectors (such as pPIC9, pPIC9K, pPICZα, etc.), in which the secretory expression vector usually contains a signal peptide sequence inserted behind the promoter. The characteristics of the expression vectors commonly used at present are summarized in detail in a recent report [28].
Integrative plasmids are frequently used for exogenous gene expression in P. pastoris. Such vectors are typically constructed as E. coli/P. pastoris shuttle vectors. On the one hand, these vectors contain elements for plasmid amplification in E. coli, including an origin of replication and a selection marker, which is usually antibiotic resistance. On the other hand, these vectors carry components required for heterologous gene expression in P. pastoris, including the promoter/terminator, the multiple cloning site, and a proper selection marker. The selection marker can be an auxotrophic marker (HIS4, ARG4, URA3, etc.) or an antibiotic resistance marker (zeocin, G418, etc.).
To introduce a foreign gene into P. pastoris, episomal vectors are generally accompanied with easier manipulation and higher efficiency compared with gene integration into the chromosome. However, natural autoreplicative vectors have limited applications in P. pastoris due to their instability and the uneven distribution among progeny cells during cell division. To solve these issues, the genome of P. pastoris has been analyzed and the chromosome-2 centromeric DNA sequence has been identified to facilitate stable autoreplication and accurate distribution of the plasmid [29]. The stability of the episomal vectors can also be improved by introducing the autonomously replicating sequences (ARS) from other organisms, such as the panARS derived from Kluyveromyces lactis [30].
3.3. Promoters
3.3.1. AOX1 Promoter
Upon entry into P. pastoris, methanol is decomposed into formaldehyde and hydrogen peroxide by alcohol oxidase encoded by AOX1 and AOX2. However, the enzyme activity is mostly provided by AOX1 due to the low expression level of AOX2 [21]. The endogenous promoter controlling AOX1 expression, i.e., PAOX1 upstream of AOX1, can be efficiently induced by methanol at low concentrations, with the optimal concentration ranging from 0.5% to 2.0% [6].
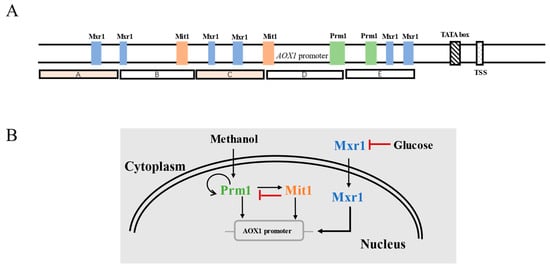
In recent years, with the continuous mechanistic research on the transcriptional regulation of PAOX1, the cis-acting elements and transcription factors that affect the transcriptional activity of this promoter have been discovered. Inan et al. divided PAOX1 into five segments (A~E), of which fragments B and E play a promotional role for AOX1 expression (Figure 3A) [31]. In P. pastoris, the response of PAOX1 to methanol is positively regulated by a cascade of transcription factors including Mit1, Mxr1, and Prm1, which bind to different sites of PAOX1 [32]. Among them, Mxr1 is closely related to the carbon-source-induced repression of PAOX1. When methanol is used as the carbon source, Mxr1 is transferred from the cytoplasm to the nucleus, leading to the derepression of PAOX1. At this point, Prm1 activates its own expression and the expression of Mit1, thus inducing strong activation of PAOX1. The strength of PAOX1 can be improved via Mit1 overexpression [33]; however, Mit1 exerts feedback inhibition of Prm1 expression (Figure 3B) [34]. In consequence, the expression level of Mit1 needs to be delicately tuned to achieve the optimal expression of genes controlled by PAOX1.

Figure 3. (A) The structure of PAOX1. (B) Transcriptional regulation of PAOX1.
3.3.2. GAP Promoter
Inducible promoters generally have some intrinsic disadvantages, such as costs and toxicity associated with the inducers and concerns of leaky expression of the target genes. A preferred alternative is the constitutive promoter. Genes regulated by constitutive promoters can be normally expressed during cell growth without the involvement of inducers. In P. pastoris, the most commonly used constitutive promoter is the glyceraldehyde-3-phosphate dehydrogenase promoter (PGAP), which is considered as a standard promoter for methanol-free expression systems [35][36]. This promoter is regulated by the metabolism of carbon sources, and its transcription level is the highest in the presence of glucose and is the lowest when cells are fed with methanol [37].
PGAP is generally weaker than PAOX1 and attempts have hence been made to improve the strength of PGAP. Ata et al. [38] analyzed the transcription factor binding sites of PGAP. Via targeted deletion or overexpression of these sites, a promoter library was constructed with different strength in initiating gene expression.
3.3.3. Other Promoters
Although the use of methanol as the sole source of carbon and energy is a major advantage of the P. pastoris cell factories, the toxicity and safety issues of methanol impose many limitations on practical industrial applications [39]. To solve this problem, methanol-free induction systems have been developed based on PAOX1. Chang et al. synthesized a positive feedback circuit in which PAOX2-driven Mxr1 promotes the transcription of PAOX1, while PAOX1 is induced under glycerol starvation or in the absence of carbon sources [40]. Kinases have been proposed as potential targets for regulating the repression of PAOX1 in the presence of a common carbon source such as glycerol [41]. By targeting the genes encoding glycerol kinase (gut1) and dihydroxyacetone kinase (dak), non-methanol-inducible PAOX1 expression systems were constructed using glycerol and dihydroxyacetone as carbon sources to induce PAOX1 expression, respectively, although the induction was not as efficient as that induced by methanol [42].
3.3.4. Synthetic Core Promoter Engineering
The core promoter region plays a pivotal role in the regulation of gene expression, and its genetic modification is an important content of promoter engineering. The core promoter is the region necessary for RNA polymerase to recognize and initiate transcription, and it consists of the RNA polymerase binding site, the TATA box, and the transcription start site. In P. pastoris, fully synthetic core promoters and the 5′-untranslated region have been designed and applied to PAOX1, resulting in a series of promoter libraries with different expression levels [43].
3.4. Signal Peptides
Secretory expression of proteins is generally dependent on cleavable signal peptides (usually 15–50 amino acids) that direct the transmembrane transfer of the newly synthesized peptides and proteins. These signal peptides, mostly located at the N-termini of the secreted proteins, usually contain three domains, i.e., the positively charged basic N-terminus (1–5 amino acids), the hydrophobic center that forms a helical structure (7–15 amino acids), and the highly polar C-terminus (3–7 amino acids) that serves as the cleavage site [44]. With a great impact on the extent of protein folding and the rate of protein secretion, signal peptides play a crucial role in high-level expression and secretion of functional proteins [45].
The signal peptides commonly used for protein secretion in P. pastoris include the signal sequence of α-factor and invertase-2 (SUC2) of S. cerevisiae, and P. pastoris acid phosphatase signal peptide (PHO1). Among these, α-factor is used most frequently and is mainly suitable for the secretory expression of peptides and small proteins. To further improve the efficiency of secretion, this signal peptide has been engineered through codon optimization, modification of the hydrophobic region, addition of spacer sequences, and site-directed mutagenesis [46][47][48].
3.5. CRISPR/Cas9 Genome Editing in Pichia pastoris
Since P. pastoris is an important workhorse for the synthesis of various bio-products, it is crucial to establish efficient and concise gene editing technologies for the genetic modifications of this microbe. Traditionally, gene insertion/deletion/replacement of P. pastoris relies on homologous recombination, which is inefficient with a low rate of success even when long homologous arms (sometimes more than 1 kb) are used. This is due to the domination of NHEJ over homologous recombination in this yeast [11]. Deletion of KU70 impairs NHEJ and significantly facilitates homologous recombination at the expense of a lower transformation efficiency and slower cell growth.
CRISPR/Cas9 introduces breaks in DNA sequences complementary to the sgRNA, which are then repaired by host cells. Thereby, genetic modifications can be introduced programmably at desired locations by using sgRNA with particularly designed sequences. Compared with traditional homologous recombination-guided genomic modification, CRISPR/Cas9 is highly flexible in the sense that only sgRNA needs to be re-designed for each independent genomic modification process. As one of the most potent and convenient gene editing technologies, CRISPR/Cas9 has been explored extensively in the engineering of P. pastoris [8][49][50].
CRISPR/Cas9-mediated genomic editing in P. pastoris relies on the correct expression of Cas9 and sgRNA in the nucleus, which can be affected by a series of factors. To improve the expression, researchers used RNA Pol II promoter for sgRNA expression, added ribozyme sequences both upstream and downstream of sgRNA, and optimized the codon of Cas9. After such optimization, near 100% efficiency could be achieved in P. pastoris for gene deletion, and multiplex gene deletion and targeted gene insertion was achieved efficiently with the aid of NHEJ [51]. This system was further introduced into a KU70-knockout strain, so that DNA breaks could be repaired through homologous recombination. Despite lower cell viability, near 100% efficiency of gene integration was achieved [52].
The biosynthetic pathways of value-added compounds (natural products and bulk chemicals) are often complex and involve multiple pathway genes. Therefore, a competent genetic tool that can manipulate multi-gene pathways has important implications for the application of P. pastoris as a cell factory. Based on the KU70 knockout strain, a CRISPR/Cas9-mediated marker-less multi-site gene integration method has been developed, for which various sgRNA targets are designed within 100 bp upstream of the promoter or downstream of the terminator. Using this method, the integration efficiency of double-locus could reach 57.7%–70% [53]. The same method was used to establish a standardized CRISPR-based synthetic biology toolkit, in which the integration efficiency of double-locus could reach ~93% [54]. This toolkit allowed for one-step assembly of the biosynthetic pathways of 2,3-butanediol, β-carotene, zeaxanthin, and astaxanthin [54].
4. Adaptive Laboratory Evolution of Pichia pastoris
Adaptive laboratory evolution (ALE) is a method to artificially simulate the mutation and selection process in natural evolution under laboratory conditions such that the directed evolution of microorganisms can be achieved within a short period of time and mutated microbes with desired traits can be screened [55]. Compared with metabolic engineering, ALE only focuses on the generation of appropriate interference factors without detailed information on the intricate and intersecting metabolic networks, thus demonstrating broad applicability and strong practicability. ALE is one of the most effective methods of strain construction toward high-level synthesis of bio-products [56][57]. Although widely used in S. cerevisiae and E. coli, ALE only has limited applications in P. pastoris, and there is still a large space for development.
Efficient use of carbon sources and substrates is key to the high-level microbial production of bio-products, and ALE has been adopted in promoting the metabolic performances of P. pastoris on various nutrients or substrates. Moser et al. investigated the effect of growth media on cell growth and recombinant protein production in P. pastoris X-33 using methanol as a carbon source for continuous subculture in eutrophic medium YPM and low-nutrient medium BMM. After approximately 250 generations, evolved strains showed higher growth rates. Whole genome sequencing identified mutations in the AOX1 gene involving the methanol binding region and its vicinity, leading to, surprisingly, a decline in AOX activity, possibly due to less intracellular accumulation of the toxic compound formaldehyde. Such methanol adaptation led to significantly higher titers of recombinant human serum albumin and fused lobes hexosaminidases [58].
5. Practical Applications of Pichia pastoris as a Cell Factory
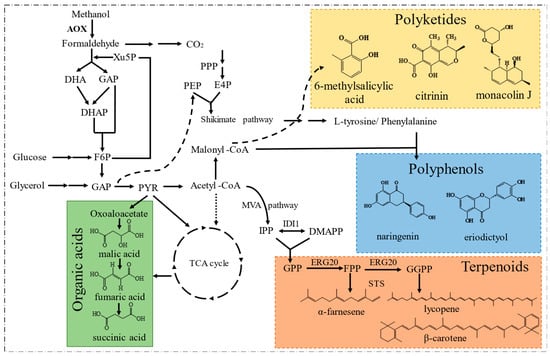
Since Philips Petroleum Company released the P. pastoris expression system to academic research laboratories in 1993, the expression system has developed rapidly [59]. P. pastoris has gradually replaced S. cerevisiae as the eukaryotic expression system because it secretes very few endogenous proteins and has glycosylation similar to that in mammalian cells. P. pastoris has thus been gradually developed into a common host for the expression of medical and industrial enzymes, and thousands of recombinant proteins have been successfully produced [60]. In addition, natural products with diverse structures have also been synthesized in this host (Figure 4), which overcomes the disadvantages associated with chemical synthesis or extraction from plants that are traditionally used for their production. These accomplishments have promoted the engineering and development of P. pastoris as a potent and potential microbial platform [8].

Figure 4. The portfolio of typical compounds produced by engineered P. pastoris.
5.1. Recombinant Proteins
5.1.1. Nanobodies
Nanobodies, the natural antibodies first found in the serum of camels and sharks, are the smallest units known to bind antigens [61][62]. Compared with traditional antibodies, nanobodies have unique properties such as strong antigen binding, low immunogenicity, high solubility and stability, and low molecular weight, which offer potential advantages in disease diagnosis and treatment [63]. For example, nanobody neutralization therapy has been employed in the treatment of the coronavirus COVID-19 [64]. Nanobodies can be stably expressed in P. pastoris besides prokaryotic hosts [65].
5.1.2. Human Proteins
Human serum albumin (HSA) is the major protein in human plasma and is widely used for drug delivery. Its recombinant expression in P. pastoris GS115 has been reported, and a high yield of 8.86 g/L was obtained after process optimization [66]. Human coagulation factor XII plays an important role in thrombosis, and its abundant supply is necessary for inhibitor screening in the development of antithrombotic drugs. The recombinant serine protease domain of human coagulation factor XII has been expressed in P. pastoris X-33 with a yield of 20 mg/L and a clotting activity similar to that of its natural counterpart [67]. Compared with these cellular proteins which can be expressed easily in the soluble form, high-level microbial expression of human membrane proteins can be a real challenge. Nonetheless, a human multichannel membrane protein named sterol ∆8-∆7 isomerase has been successfully expressed in the form of a GFP fusion in P. pastoris as well as in E. coli and S. cerevisiae, with the best expression achieved in P. pastoris at 200 mg/L in shake flasks and 1000 mg/L in condensed culture [68].
5.2. Value-Added Compounds
5.2.1. Terpenoids
Terpenoids are secondary metabolites with isoprene as the basic structural unit, and they mainly include monoterpenoids, sesquiterpenoids, diterpenoids, triterpenoids, and polyterpenoids [69]. Most of these compounds have anti-tumor, anti-inflammatory, and immunomodulatory effects, and are widely used in food processing and pharmaceutical manufacturing industries [70]. Yeast cells generally produce more precursors than bacteria for terpenoid biosynthesis, including DMAPP (dimethylallyl diphosphate) and IPP (isopentenyl diphosphate) [71]. Compared with other yeast chasses, P. pastoris can reach a higher fermentation density, produce fewer metabolic byproducts, and present strong tolerance to complex environments. These traits make P. pastoris suitable for terpenoid biosynthesis.
5.2.2. Polysaccharides
Polysaccharides widely exist in nature and participate in important cellular processes as an energy source and organizational structure. Polysaccharides can be used as efficient drug carriers due to their biodegradability, biocompatibility, and generally low costs [72]. Polysaccharide biosynthesis using engineered microorganisms has been attracting increasing attention, with some of the focus laid on P. pastoris due to its abundant supply of sugar precursors.
5.2.3. Polyketides
Polyketides are a class of widely distributed natural products produced during the secondary metabolism of plants or microorganisms, with rich chemical properties and unique physiological activities. Polyketide biosynthesis in P. pastoris was first reported in 2013 for the fungal polyketide compound 6-methylsalicylic acid (6-MSA) [73]. The genes encoding Aspergillus nidulans phosphopantetheinyl transferase (npgA) and Aspergillus terrus 6-MSA synthase (atX) were integrated into the genome of P. pastoris via homologous recombination, thus producing 2.2 g/L of 6-MSA in a 5 L bioreactor upon 20 h of methanol induction. On this basis, a more structurally complex polyketide citrinin was also synthesized in P. pastoris by assembling npgA, Monascus purpureus citrinin polyketide synthase gene pksCT, and several genes in the citrinin gene cluster [74].
6. Conclusions
Pichia pastoris has been developed to produce proteins for four decades and has emerged as a new chassis to produce diverse chemicals and natural products in recent years. With the unique property of utilizing methanol as a sole carbon source, metabolic engineering of P. pastoris is attracting increasing attention amid the great concern on global energy security, given the potential of methanol as a supply of energy and carbon source for biomanufacturing. P. pastoris has shown great success in the biosynthesis of some natural products with the titers reaching >1 g/L, indicating very promising applications for industrialization and commercialization.
References
- Kurtzman, C.P. Biotechnological strains of Komagataella (Pichia) pastoris are Komagataella phaffii as determined from multigene sequence analysis. J. Ind. Microbiol. Biotechnol. 2009, 36, 1435–1438.
- Kulagina, N.; Besseau, S.; Godon, C.; Goldman, G.H.; Papon, N.; Courdavault, V. Yeasts as biopharmaceutical production platforms. Front. Fungal Biol. 2021, 2, 733492.
- Cereghino, G.P.L.; Cereghino, J.L.; Ilgen, C.; Cregg, J.M. Production of recombinant proteins in fermenter cultures of the yeast Pichia pastoris. Curr. Opin. Biotechnol. 2002, 13, 329–332.
- Cereghino, J.L.; Cregg, J.M. Heterologous protein expression in the methylotrophic yeast Pichia pastoris. FEMS Microbiol. Rev. 2000, 24, 45–66.
- Hasslacher, M.; Schall, M.; Hayn, M.; Bona, R.; Rumbold, K.; Lückl, J.; Griengl, H.; Kohlwein, S.D.; Schwab, H. High-level intracellular expression of hydroxynitrile lyase from the tropical rubber tree Hevea brasiliensis in microbial hosts. Protein Expr. Purif. 1997, 11, 61–71.
- Karbalaei, M.; Rezaee, S.A.; Farsiani, H. Pichia pastoris: A highly successful expression system for optimal synthesis of heterologous proteins. J. Cell Physiol. 2020, 235, 5867–5881.
- Pan, Y.; Yang, J.; Wu, J.; Yang, L.; Fang, H. Current advances of Pichia pastoris as cell factories for production of recombinant proteins. Front. Microbiol. 2022, 13, 1059777.
- Gao, J.; Jiang, L.; Lian, J. Development of synthetic biology tools to engineer Pichia pastoris as a chassis for the production of natural products. Synth. Syst. Biotechnol. 2021, 6, 110–119.
- Grinna, L.S.; Tschopp, J.F. Size distribution and general structural features of N-linked oligosaccharides from the methylotrophic yeast, Pichia pastoris. Yeast 1989, 5, 107–115.
- Zhu, T.; Zhao, T.; Bankefa, O.E.; Li, Y. Engineering unnatural methylotrophic cell factories for methanol-based biomanufacturing: Challenges and opportunities. Biotechnol. Adv. 2020, 39, 107467.
- Cai, P.; Duan, X.; Wu, X.; Gao, L.; Ye, M.; Zhou, Y.J. Recombination machinery engineering facilitates metabolic engineering of the industrial yeast Pichia pastoris. Nucleic Acids Res. 2021, 49, 7791–7805.
- Batt, C.A. Pichia pastoris. In Encyclopedia of Food Microbiology; Academic Press: London, UK, 2014; pp. 42–46.
- Heistinger, L.; Moser, J.; Tatto, N.E.; Valli, M.; Gasser, B.; Mattanovich, D. Identification and characterization of the Komagataella phaffii mating pheromone genes. FEMS Yeast Res. 2018, 18, foy051.
- Dragosits, M.; Stadlmann, J.; Albiol, J.; Baumann, K.; Maurer, M.; Gasser, B.; Sauer, M.; Altmann, F.; Ferrer, P.; Mattanovich, D. The effect of temperature on the proteome of recombinant Pichia pastoris. J. Proteome Res. 2009, 8, 1380–1392.
- Rosenbergová, Z.; Kántorová, K.; Šimkovič, M.; Breier, A.; Rebroš, M. Optimisation of recombinant myrosinase production in Pichia pastoris. Int. J. Mol. Sci. 2021, 22, 3677.
- Ata, Ö.; Rebnegger, C.; Tatto, N.E.; Valli, M.; Mairinger, T.; Hann, S.; Steiger, M.G.; Çalık, P.; Mattanovich, D. A single Gal4-like transcription factor activates the Crabtree effect in Komagataella phaffii. Nat. Commun. 2018, 9, 4911.
- Zhan, C.; Wang, S.; Sun, Y.; Dai, X.; Liu, X.; Harvey, L.; McNeil, B.; Yang, Y.; Bai, Z. The Pichia pastoris transmembrane protein GT1 is a glycerol transporter and relieves the repression of glycerol on AOX1 expression. FEMS Yeast Res. 2016, 16, fow033.
- Liu, B.; Zhang, Y.; Zhang, X.; Yan, C.; Zhang, Y.; Xu, X.; Zhang, W. Discovery of a rhamnose utilization pathway and rhamnose-inducible promoters in Pichia pastoris. Sci. Rep. 2016, 6, 27352.
- Liu, B.; Li, H.; Zhou, H.; Zhang, J. Enhancing xylanase expression by Komagataella phaffii by formate as carbon source and inducer. Appl. Microbiol. Biotechnol. 2022, 106, 7819–7829.
- Gabaldón, T. Peroxisome diversity and evolution. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 765–773.
- Vogl, T.; Glieder, A. Regulation of Pichia pastoris promoters and its consequences for protein production. New Biotechnol. 2013, 30, 385–404.
- Yu, X.-W.; Lu, X.; Zhao, L.-S.; Xu, Y. Impact of NH4+ nitrogen source on the production of Rhizopus oryzae lipase in Pichia pastoris. Process Biochem. 2013, 48, 1462–1468.
- Rumjantsev, A.M.; Bondareva, O.V.; Padkina, M.V.; Sambuk, E.V. Effect of nitrogen source and inorganic phosphate concentration on methanol utilization and PEX genes expression in Pichia pastoris. Sci. World J. 2014, 2014, 743615.
- Ayed, A.; Rabhi, I.; Dellagi, K.; Kallel, H. High level production and purification of human interferon α2b in high cell density culture of Pichia pastoris. Enzym. Microb. Technol. 2008, 42, 173–180.
- Ahmad, M.; Hirz, M.; Pichler, H.; Schwab, H. Protein expression in Pichia pastoris: Recent achievements and perspectives for heterologous protein production. Appl. Microbiol. Biotechnol. 2014, 98, 5301–5317.
- Barone, G.D.; Emmerstorfer-Augustin, A.; Biundo, A.; Pisano, I.; Coccetti, P.; Mapelli, V.; Camattari, A. Industrial production of proteins with Pichia pastoris-Komagataella phaffii. Biomolecules 2023, 13, 441.
- Gleeson, M.A.; White, C.E.; Meininger, D.P.; Komives, E.A. Generation of protease-deficient strains and their use in heterologous protein expression. Methods Mol. Biol. 1998, 103, 81–94.
- Vijayakumar, V.E.; Venkataraman, K. A Systematic review of the potential of Pichia pastoris (Komagataella phaffii) as an alternative host for biologics production. Mol. Biotechnol. 2023, 1–19.
- Nakamura, Y.; Nishi, T.; Noguchi, R.; Ito, Y.; Watanabe, T.; Nishiyama, T.; Aikawa, S.; Hasunuma, T.; Ishii, J.; Okubo, Y.; et al. A Stable, Autonomously Replicating Plasmid Vector C containing Pichia pastoris centromeric DNA. Appl. Envion. Microbiol. 2018, 84, e02882-17.
- Gu, Y.; Gao, J.; Cao, M.; Dong, C.; Lian, J.; Huang, L.; Cai, J.; Xu, Z. Construction of a series of episomal plasmids and their application in the development of an efficient CRISPR/Cas9 system in Pichia pastoris. World J. Microbiol. Biotechnol. 2019, 35, 79.
- Inan, M. Studies on the Alcohol Oxidase (AOX1) Promoter of Pichia pastoris; The University of Nebraska: Lincoln, NE, USA, 2000.
- Wang, X.; Wang, Q.; Wang, J.; Bai, P.; Shi, L.; Shen, W.; Zhou, M.; Zhou, X.; Zhang, Y.; Cai, M. Mit1 transcription factor mediates methanol signaling and regulates the Alcohol Oxidase 1 (AOX1) Promoter in Pichia pastoris. J. Biol. Chem. 2016, 291, 6245–6261.
- Haghighi Poodeh, S.; Ranaei Siadat, S.O.; Arjmand, S.; Khalifeh Soltani, M. Improving AOX1 promoter efficiency by overexpression of Mit1 transcription factor. Mol. Biol. Rep. 2022, 49, 9379–9386.
- Sahu, U.; Krishna Rao, K.; Rangarajan, P.N. Trm1p, a Zn(II)2Cys6-type transcription factor, is essential for the transcriptional activation of genes of methanol utilization pathway, in Pichia pastoris. Biochem. Biophys. Res. Commun. 2014, 451, 158–164.
- Waterham, H.R.; Digan, M.E.; Koutz, P.J.; Lair, S.V.; Cregg, J.M. Isolation of the Pichia pastoris glyceraldehyde-3-phosphate dehydrogenase gene and regulation and use of its promoter. Gene 1997, 186, 37–44.
- Garrigós-Martínez, J.; Vuoristo, K.; Nieto-Taype, M.A.; Tähtiharju, J.; Uusitalo, J.; Tukiainen, P.; Schmid, C.; Tolstorukov, I.; Madden, K.; Penttilä, M.; et al. Bioprocess performance analysis of novel methanol-independent promoters for recombinant protein production with Pichia pastoris. Microb. Cell Factories 2021, 20, 74.
- Baghban, R.; Farajnia, S.; Ghasemi, Y.; Mortazavi, M.; Zarghami, N.; Samadi, N. New developments in Pichia pastoris expression system, review and update. Curr. Pharm. Biotechnol. 2018, 19, 451–467.
- Ata, Ö.; Prielhofer, R.; Gasser, B.; Mattanovich, D.; Çalık, P. Transcriptional engineering of the glyceraldehyde-3-phosphate dehydrogenase promoter for improved heterologous protein production in Pichia pastoris. Biotechnol. Bioeng. 2017, 114, 2319–2327.
- Wang, J.; Wang, X.; Shi, L.; Qi, F.; Zhang, P.; Zhang, Y.; Zhou, X.; Song, Z.; Cai, M. Methanol-independent protein expression by AOX1 Promoter with trans-acting elements engineering and glucose-glycerol-shift induction in Pichia pastoris. Sci. Rep. 2017, 7, 41850.
- Chang, C.-H.; Hsiung, H.-A.; Hong, K.-L.; Huang, C.-T. Enhancing the efficiency of the Pichia pastoris AOX1 promoter via the synthetic positive feedback circuit of transcription factor Mxr1. BMC Biotechnol. 2018, 18, 81.
- Shen, W.; Kong, C.; Xue, Y.; Liu, Y.; Cai, M.; Zhang, Y.; Jiang, T.; Zhou, X.; Zhou, M. Kinase screening in Pichia pastoris identified promising targets involved in cell growth and Alcohol Oxidase 1 promoter (PAOX1) regulation. PLoS ONE 2016, 11, e0167766.
- Shen, W.; Xue, Y.; Liu, Y.; Kong, C.; Wang, X.; Huang, M.; Cai, M.; Zhou, X.; Zhang, Y.; Zhou, M. A novel methanol-free Pichia pastoris system for recombinant protein expression. Microb. Cell Factories 2016, 15, 178.
- Vogl, T.; Ruth, C.; Pitzer, J.; Kickenweiz, T.; Glieder, A. Synthetic core promoters for Pichia pastoris. ACS Synth. Biol. 2014, 3, 188–191.
- Damasceno, L.M.; Anderson, K.A.; Ritter, G.; Cregg, J.M.; Old, L.J.; Batt, C.A. Cooverexpression of chaperones for enhanced secretion of a single-chain antibody fragment in Pichia pastoris. Appl. Microbiol. Biotechnol. 2007, 74, 381–389.
- Owji, H.; Nezafat, N.; Negahdaripour, M.; Hajiebrahimi, A.; Ghasemi, Y. A comprehensive review of signal peptides: Structure, roles, and applications. Eur. J. Cell Biol. 2018, 97, 422–441.
- Ahn, J.; Jang, M.J.; Ang, K.S.; Lee, H.; Choi, E.S.; Lee, D.Y. Codon optimization of Saccharomyces cerevisiae mating factor alpha prepro-leader to improve recombinant protein production in Pichia pastoris. Biotechnol. Lett. 2016, 38, 2137–2143.
- Barrero, J.J.; Casler, J.C.; Valero, F.; Ferrer, P.; Glick, B.S. An improved secretion signal enhances the secretion of model proteins from Pichia pastoris. Microb. Cell Factories 2018, 17, 161.
- Ito, Y.; Ishigami, M.; Hashiba, N.; Nakamura, Y.; Terai, G.; Hasunuma, T.; Ishii, J.; Kondo, A. Avoiding entry into intracellular protein degradation pathways by signal mutations increases protein secretion in Pichia pastoris. Microb. Biotechnol. 2022, 15, 2364–2378.
- Peña, D.A.; Gasser, B.; Zanghellini, J.; Steiger, M.G.; Mattanovich, D. Metabolic engineering of Pichia pastoris. Metab. Eng. 2018, 50, 2–15.
- Wu, X.; Cai, P.; Yao, L.; Zhou, Y.J. Genetic tools for metabolic engineering of Pichia pastoris. Eng. Microbiol. 2023, 3, 100094.
- Weninger, A.; Hatzl, A.-M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial optimization of CRISPR/Cas9 expression enables precision genome engineering in the methylotrophic yeast Pichia pastoris. J. Biotechnol. 2016, 235, 139–149.
- Weninger, A.; Fischer, J.E.; Raschmanová, H.; Kniely, C.; Vogl, T.; Glieder, A. Expanding the CRISPR/Cas9 toolkit for Pichia pastoris with efficient donor integration and alternative resistance markers. J. Cell Biochem. 2018, 119, 3183–3198.
- Liu, Q.; Shi, X.; Song, L.; Liu, H.; Zhou, X.; Wang, Q.; Zhang, Y.; Cai, M. CRISPR-Cas9-mediated genomic multiloci integration in Pichia pastoris. Microb. Cell Factories 2019, 18, 144.
- Gao, J.; Xu, J.; Zuo, Y.; Ye, C.; Jiang, L.; Feng, L.; Huang, L.; Xu, Z.; Lian, J. Synthetic biology toolkit for marker-less integration of multigene pathways into Pichia pastoris via CRISPR/Cas9. ACS Synth. Biol. 2022, 11, 623–633.
- Sandberg, T.E.; Salazar, M.J.; Weng, L.L.; Palsson, B.O.; Feist, A.M. The emergence of adaptive laboratory evolution as an efficient tool for biological discovery and industrial biotechnology. Metab. Eng. 2019, 56, 1–16.
- Hirasawa, T.; Maeda, T. Adaptive laboratory evolution of microorganisms: Methodology and application for bioproduction. Microorganisms 2022, 11, 92.
- Lee, S.; Kim, P. Current status and applications of adaptive laboratory evolution in industrial microorganisms. J. Microbiol. Biotechnol. 2020, 30, 793–803.
- Moser, J.W.; Prielhofer, R.; Gerner, S.M.; Graf, A.B.; Wilson, I.B.H.; Mattanovich, D.; Dragosits, M. Implications of evolutionary engineering for growth and recombinant protein production in methanol-based growth media in the yeast Pichia pastoris. Microb. Cell Factories 2017, 16, 49.
- Higgins, D.R.; Cregg, J.M. Introduction to Pichia pastoris. Methods Mol. Biol. 1998, 103, 1–15.
- Damasceno, L.M.; Huang, C., Jr.; Batt, C.A. Protein secretion in Pichia pastoris and advances in protein production. Appl. Microbiol. Biotechnol. 2012, 93, 31–39.
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448.
- Greenberg, A.S.; Avila, D.; Hughes, M.; Hughes, A.; McKinney, E.C.; Flajnik, M.F. A new antigen receptor gene family that undergoes rearrangement and extensive somatic diversification in sharks. Nature 1995, 374, 168–173.
- Hassanzadeh-Ghassabeh, G.; Devoogdt, N.; De Pauw, P.; Vincke, C.; Muyldermans, S. Nanobodies and their potential applications. Nanomedicine 2013, 8, 1013–1026.
- Huo, J.; Mikolajek, H.; Le Bas, A.; Clark, J.J.; Sharma, P.; Kipar, A.; Dormon, J.; Norman, C.; Weckener, M.; Clare, D.K.; et al. A potent SARS-CoV-2 neutralising nanobody shows therapeutic efficacy in the Syrian golden hamster model of COVID-19. Nat. Commun. 2021, 12, 5469.
- Muyldermans, S. A guide to: Generation and design of nanobodies. FEBS J. 2021, 288, 2084–2102.
- Zhu, W.; Gong, G.; Pan, J.; Han, S.; Zhang, W.; Hu, Y.; Xie, L. High level expression and purification of recombinant human serum albumin in Pichia pastoris. Protein Expr. Purif. 2018, 147, 61–68.
- Peng, B.; Xue, G.; Xu, D.; Feng, Z.; Chen, J.; Huang, M.; Lu, H.; Gong, L. Expression and purification of recombinant serine protease domain of human coagulation factor XII in Pichia pastoris. Biosci. Biotechnol. Biochem. 2019, 83, 1815–1821.
- Cai, H.; Yao, H.; Li, T.; Tang, Y.; Li, D. High-level heterologous expression of the human transmembrane sterol Δ8,Δ7-isomerase in Pichia pastoris. Protein Expr. Purif. 2019, 164, 105463.
- Tholl, D. Biosynthesis and biological functions of terpenoids in plants. Adv. Biochem. Eng. Biotechnol. 2015, 148, 63–106.
- Akazawa, H.; Kohno, H.; Tokuda, H.; Suzuki, N.; Yasukawa, K.; Kimura, Y.; Manosroi, A.; Manosroi, J.; Akihisa, T. Anti-inflammatory and anti-tumor-promoting effects of 5-deprenyllupulonol C and other compounds from Hop (Humulus lupulus L.). Chem. Biodivers. 2012, 9, 1045–1054.
- Zhang, Y.; Nielsen, J.; Liu, Z. Engineering yeast metabolism for production of terpenoids for use as perfume ingredients, pharmaceuticals and biofuels. FEMS Yeast Res. 2017, 17, fox080.
- Prateeksha; Sharma, V.K.; Liu, X.; Oyarzún, D.A.; Abdel-Azeem, A.M.; Atanasov, A.G.; Hesham, A.E.-L.; Barik, S.K.; Gupta, V.K.; Singh, B.N. Microbial polysaccharides: An emerging family of natural biomaterials for cancer therapy and diagnostics. Semin. Cancer Biol. 2022, 86, 706–731.
- Gao, L.; Cai, M.; Shen, W.; Xiao, S.; Zhou, X.; Zhang, Y. Engineered fungal polyketide biosynthesis in Pichia pastoris: A potential excellent host for polyketide production. Microb. Cell Factories 2013, 12, 77.
- Xue, Y.; Kong, C.; Shen, W.; Bai, C.; Ren, Y.; Zhou, X.; Zhang, Y.; Cai, M. Methylotrophic yeast Pichia pastoris as a chassis organism for polyketide synthesis via the full citrinin biosynthetic pathway. J. Biotechnol. 2017, 242, 64–72.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
19 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




