Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Claudia Benavente | -- | 3352 | 2024-01-11 23:01:54 | | | |
| 2 | Mona Zou | Meta information modification | 3352 | 2024-01-15 09:51:26 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Chen, C.C.; Benavente, C.A. Exosomal Cargos on Osteosarcoma Progression. Encyclopedia. Available online: https://encyclopedia.pub/entry/53766 (accessed on 07 February 2026).
Chen CC, Benavente CA. Exosomal Cargos on Osteosarcoma Progression. Encyclopedia. Available at: https://encyclopedia.pub/entry/53766. Accessed February 07, 2026.
Chen, Claire C., Claudia A. Benavente. "Exosomal Cargos on Osteosarcoma Progression" Encyclopedia, https://encyclopedia.pub/entry/53766 (accessed February 07, 2026).
Chen, C.C., & Benavente, C.A. (2024, January 11). Exosomal Cargos on Osteosarcoma Progression. In Encyclopedia. https://encyclopedia.pub/entry/53766
Chen, Claire C. and Claudia A. Benavente. "Exosomal Cargos on Osteosarcoma Progression." Encyclopedia. Web. 11 January, 2024.
Copy Citation
Osteosarcoma (OS) is a primary malignant bone tumor with high metastasis. Poor prognosis highlights a clinical need for novel therapeutic strategies. Exosomes, also known as extracellular vesicles, have been identified as essential players in the modulation of cancer. OS-derived exosomes can drive pro-tumorigenic or anti-tumorigenic phenotypes by transferring specific cargos, including proteins, nucleic acids, and metabolites, to neighboring cells, significantly impacting the regulation of cellular processes.
exosomes
extracellular vesicles
osteosarcoma
1. Exosomal Cargos Mediate Metastasis of Osteosarcoma
Substantial in vitro and in vivo evidence strongly supports the important role and potential clinical relevance of exosomal cargo in both the development and progression of OS. In a recent study, Jerez et al. investigated exosomal cargos from human OS (SAOS2, MG63, U2OS, HOS, and 143B) or osteoblast cell lines (hFOB 1.19 cells) with different metastatic potential. Using miRNA sequencing profiling, the researchers identified four miRNAs (miR-21-5p, miR-143-3p, miR-148a-3p, and 181a-5p) that were significantly enriched, with higher expression levels in metastatic SAOS2 cells compared to those of nonmetastatic MG63 cells. Gene ontology analysis suggests these miRNAs may regulate cell adhesion and apoptosis [1]. However, no phenotypic validation was included in this study. Support for some of these observations comes from other independent studies and are contradicted by others. Indeed, miR-21-5p is considered an oncomir in solid and hematological malignancies [2][3][4], where exosomal miR-21-5p mediates crosstalk within the tumor microenvironment (TME) to prepare the metastatic niche. Inhibition of miR-21-5p leads to decreased proliferation and metastasis of OS by targeting PTEN and modulating the TGF-β1 signaling pathway [1][5][6][7][8]. OS-derived exosomal miR-1307 regulates AGAP1 and induces OS tumorigenesis [9]. In vitro and in vivo studies have demonstrated that exosomes secreted by OS, carrying miR-195-3p, enhance the proliferation and invasion of 143B cells [10]. Gong et al. demonstrated that miR-675 is significantly upregulated in exosomes derived from metastatic OS cells in comparison to nonmetastatic OS cells, leading to enhanced migration and invasion of osteoblasts through the downregulation of calneuron 1 (CALN1) expression [11]. Exosomes released by cancer-associated fibroblasts transfer miR-1228 to OS cells, leading to the promotion of OS migration and invasion through the inhibition of endogenous suppressor of cancer cell invasion (SCAI) expression [12]. The transfer of exosomal miR-208a obtained from bone marrow mesenchymal stem cells (BMSCs) to OS cells was observed to promote OS cell proliferation, migration, and clonogenicity while inhibiting apoptosis. This effect was attributed to the downregulation of programmed cell death protein 4 (PDCD4) and activation of the Hippo and ERK1/2 signaling pathways [13]. Likewise, exosomal miR-769-5p was identified to promote OS cell proliferation and metastasis in vitro and in vivo by downregulating Dual-specific phosphatase 16 (DUSP16) and activating the JNK/p38 MAPK (mitogen-activated protein kinase) signaling pathway [14]. Upregulation of oncogenic miR-25-3p from OS-derived exosomes enhances invasion and proliferation of umbilical vein endothelial cells by targeting Dickkopf WNT signaling pathway inhibitor 3 (DKK3) [15]. Interestingly, studies on exosomal miR-143 on OS progression show contradictive results. While next-generation sequencing analysis revealed that exosomal miR-143 from OS cells is expressed at higher levels in metastatic SAOS2 cells [1], other studies suggest miR-143 could suppress tumor metastatic potential by targeting mitogen-activated protein kinase 7 (MAPK7) [16][17]. A study by Li et al. reported that up-regulation of miR-143 could dampen OS cell migration, proliferation, and invasive capability and induce apoptosis via caspase3 activation by targeting Bcl-2 [18]. Osaki et al. observed that administration of miR-143 suppresses spontaneous lung metastasis of OS cells in mice by targeting MMP-13 [19]. These apparently contradictive results can be easily reconciled considering that SAOS2 cells are low-metastatic cells [20].
2. Osteosarcoma-Derived Exosomal Cargos Mediated Immune Regulation
The interplay between tumor and immune cells within the TME contributes to OS progression and metastasis. The immune microenvironment highlights the complex crosstalk network influencing disease outcomes and therapeutic responses. Exosomes from OS cells can release growth factors, cytokines, and chemokines to facilitate intracellular communication and reprogram recipient immune cells [5]. Previous studies reveal a distinct difference in exosomal cargos altering the immune microenvironment in OS. For example, Troyer and colleagues reported exosomes carrying immunosuppressive protein cargos such as TGF-β, α fetoprotein, and heat shock proteins in OS cells compared to exosomes derived from healthy osteoblasts. They also observed that the activation and proliferation of CD4+ and CD8+ T cells were attenuated, while the expression of regulatory (FOXP3+) CD4+ was elevated, resulting in immune invasion [21]. Among the array of immune cells, tumor-associated macrophages (TAMs)—the predominant immune cells infiltrating OS—are increasingly recognized as pivotal regulators of inflammation, neoangiogenic process, and evasion of immune surveillance [22][23][24]. The dysregulation in the M1-M2 polarization of macrophages, with M1 exhibiting an anti-tumor phenotype and M2 promoting tumor-related phenotypes, governs the progression of cancer pathogenesis [25]. A recent study by Wolf-Dennen et al. demonstrated that exosomes secreted from metastatic OS cells induce macrophage M2 polarization by upregulating the expression of M2-associated cytokines and chemokines, including IL-10, TGFβ2, and CCL2. This process leads to an establishment of immunosuppressive TME and regulates phagocytosis and macrophage-mediated tumor killing [26].
Moreover, it is observed that exosomes derived from OS cells transport cargo T cell immunoglobulin and mucin domain 3 (TIM-3) to promote TAM differentiation to M2 phenotype, which facilitates OS migration, invasion, EMT and distinct metastasis in vitro and in vivo [27]. Similarly, the exosomal lncRNA ELFN1-AS1 derived from OS cells has been shown to transfer to macrophages, where it modulates M2 polarization through sponging miR-138-5p and miR-1291 and subsequently promoting tumor growth [28]. An additional exosomal cargo, Rab22a-NeoF1 fusion protein, has been reported to regulate the activation of signal transducer and activator of transcription 3 (STAT3), leading to M2 macrophage polarization and facilitation of pulmonary metastasis [29]. Likewise, Liu and colleagues demonstrated that the connection between OS malignant phenotypes and M2 polarization of TAMs is controlled by exosomal miR-221-3p, where miR-221-3p released from M2-polarized TAMs exosomes could augment tumor growth while reducing apoptosis through SOCS3/JAK2/STAT3 signaling [30].
Given that exosomes mediate crosstalk within the TME, the role of exosomes secreted by OS in immune regulation has been explored. Strikingly, recent evidence has highlighted cancer cell-derived exosomes carrying tumor-associated antigens and immunosuppressive cargos, such as FasL, PD-L1, and TGFβ, in facilitating tumor immune evasion [21][31][32]. In particular, Zhang et al. investigated the role of exosomal cargo PD-L1 in immune surveillance. Their study revealed that OS-derived exosomes carrying PD-L1 cargo could dampen T-cell activity and mediate tumor growth in vitro and in vivo [33]. These exosomal cargos have an indirect impact on promoting immune escape.
Altogether, there is a clear interplay between exosomal cargo and the immune microenvironment in OS. Modulating the release of exosomal cargo holds promise as an innovative approach to mitigate the immunosuppressive TME and improve the effectiveness of immunotherapy for OS.
3. Exosomal Cargos Mediate Osteosarcoma Drug Resistance
Neoadjuvant chemotherapy combined with surgery is the current standard of care for OS. Although chemotherapy helps reduce the tumor burden in OS patients, developing multi-drug resistance (MDR) presents a significant challenge in treating OS, resulting in metastasis and unfavorable prognosis. Previous studies have suggested exosomes mediate MDR in various types of cancer, such as glioblastoma, non-small cell lung cancer, ovarian cancer, gastric cancer, colorectal cancer, and breast cancer [34][35][36][37][38][39]. In OS, multiple studies have demonstrated that MDR arises from exosomes released from drug-resistant parental cells [40][41][42]. Torreggiani et al. demonstrated that exosomes derived from doxorubicin-resistant OS cells can be internalized by recipient cells, shuttling multi-drug resistance-associated protein 1 (MDR-1) mRNA and drug efflux pump P-glycoprotein (P-gp) to sensitive cells, thus propagating doxorubicin-resistant traits [40]. Similarly, the expression of exosomal circ_103801, originating from cisplatin (CDDP)-resistant MG63 cells, was elevated compared to the expression in MG63 cells. This transfer of circ_103801 from CDDP-resistant cells to chemosensitive OS cells. The transfer of circ_103801 from CDDP-resistant cells to chemo-sensitive OS cells was found to suppress apoptosis, reduce drug sensitivity to CDDP, increase the levels of multi-drug resistance-associated protein 1 and P-gp, and significantly contribute to the development of chemoresistance [42]. The exosomal lncRNA ANCR from doxorubicin-resistant KHOS/U2OS cells has been shown to regulate drug sensitivity to doxorubicin, as evidenced by the inability of exosomes with lncRNA ANCR knockdown to induce doxorubicin chemoresistance in mice [41]. Exosomal CCCTC-binding factor (CTCF) from CDDP-resistant OS cells activates the autophagy signaling pathway through the IGF2-AS/miR-579-3p/MSH6 axis, resulting in enhanced CDDP resistance and promoting tumor formation [43]. Given the established role of the PD-1/PD-L1 axis in promoting chemoresistance in various cancers including breast cancer, B-cell lymphoma, and small cell lung cancer, a study by Yati et al. uncovered that OS cells treated with doxorubicin can prompt the release of exosomes that mediate PD-L1 expression in OS cells through IL-1 signaling pathway [44][45][46][47]. The study suggested a relationship between chemoresistant OS-derived EVs and proinflammatory cytokine regulation. However, additional investigations, particularly those focusing on phenotypic analysis, are imperative to decipher the mechanism by which EVs regulate PD-1/PD-L1. Collectively, these findings emphasize the functional role of exosomes from chemoresistance OS cells in mediating drug resistance through mechanisms such as drug efflux, exosomal cargo delivery, and RNA transport, suggesting that targeting specific exosomal cargos could potentially help overcome MDR in OS.
4. Exosomes as Biomarkers in Osteosarcoma
Exosomes play a crucial role as carriers, shuttling various cargo molecules, including proteins, nucleic acids, and signaling molecules, contributing to the intricate process of tumorigenesis and cancer progression. To date, there is no robust biomarker available for predicting prognosis and metastasis in OS. Given the unique and complex composition of exosomes, along with their widespread presence in various tissues and biological fluids, cancer exosomes have been explored extensively as a valuable tool for biomarker applications in diagnostics, prognostic assessments, and tumor monitoring. Indeed, exosomes exhibit distinct molecular profiles between normal and pathological states, and numerous studies have discovered that exosome-derived cargos serve as potential biomarkers for OS. Exosomes derived from the blood or tissues of OS patients can be isolated, purified, and characterized by analyzing their cargos through genomics or proteomics analysis (Figure 1). The molecular signature associated with OS pathogenesis can be identified by scrutinizing the RNA or protein compositions presented in the exosomal cargo. Exosomal miRNAs have been predominantly exploited as biomarkers for the diagnosis of cancer. For example, Ye and colleagues investigated miRNA profiling in exosomes derived from plasma samples of OS patients and healthy individuals, revealing differential expression of 57 miRNAs, with 20 being upregulated and 37 downregulated. Notably, miR-92a-3p, miR-130a-3p, miR-195-3p, miR-335-5p, and let-7i-3p were significantly higher in OS patient exosomes compared to those from healthy controls [10]. In a clinical trial (NCT03108677), using plasma-derived exosomes as a liquid biopsy approach revealed significant transcriptomic alterations, including a higher tumor mutation burden, gene fusions, and aberrant gene expression in metastatic OS patients compared to primary OS. This suggests that the transcriptomic profiling of plasma exosomes holds the potential to differentiate the metastatic potential in OS patients, offering a novel approach for monitoring metastasis [48]. Gong et al. found a significantly higher expression of exosomal miR-675 in metastatic OS patients’ serums associated with prognosis [11]. Similarly, the miR-25-3p level was remarkably elevated in both the serum and tissues of OS patients and is inversely correlated with the clinical prognosis [15][49]. A separate study reported a significant decrease in the expression of plasma exosomal miR-101 in OS patients compared to healthy individuals. Notably, this decrease was even more pronounced in metastatic OS patients than in those without metastasis. This observation underscores the possibility of utilizing circulating exosomal miR-101 as a potential diagnostic biomarker for detecting OS metastasis [50]. In addition, a study by Xu et al. reported serum exosomal mRNA and miRNA profiles unveiling distinct expression patterns between OS patients exhibiting favorable and unfavorable responses to chemotherapy. Specifically, they discovered upregulation of mRNA expression in Annexin2, CDC5L, Smad2, and P27, while CIP4, MTAP, PEDF, and WWOX were found to be downregulated in OS patients with poor chemotherapeutic responses. Further, the expression levels of exosomal miR-135b, miR-148a, miR-27a, and miR-9 were elevated, whereas miR-124, miR-133a, miR-199a-3p, and miR-385 were observed to be down-regulated in OS patients with inadequate chemotherapeutic response, indicating exosomal RNA or miRNA can be used for potential biomarkers in categorizing different levels of chemotherapy sensitivity in OS [51]. Using the NGS approach, Cuscino et al. identified eight putative miRNA sequences in the exosomes derived from OS cell lines. Among these, five miRNA candidates exhibited differential expression in liquid biopsy samples from a small OS patient cohort compared to controls. These miRNA targets might represent potential biomarkers, yet further research is imperative to investigate the underlying pathological mechanisms associated with these miRNA candidates [52]. Serum-derived exosomal circ_103801 has been found to be upregulated in OS patients, and survival analysis revealed that higher circ_103801 is associated with shorter overall survival, suggesting a potential role for circ_103801 as a prognostic biomarker for OS [42]. Other exosomal non-coding RNA targets, circ_0056285, circ_0000190, and lncRNA CASC15 have shown significantly increased expression in the serum or plasma of OS patients, displaying strong diagnostic potential [53][54][55].
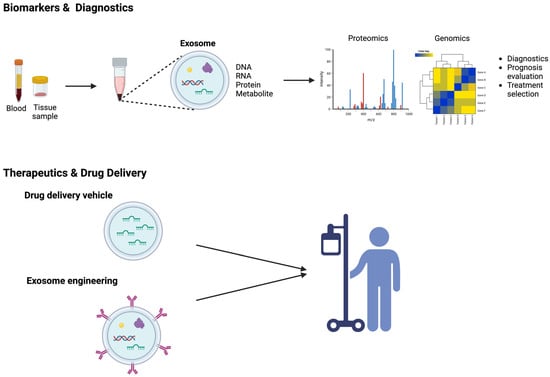
Figure 1. Exosomes as emerging biomarkers and therapeutic tools in OS. Exosomes can be extracted from body fluids or tissues, and exosomal cargos including DNAs, RNAs, proteins, and metabolites can be analyzed through proteomics or genomics to evaluate OS diagnosis, prognosis, and response to treatment. Additionally, exosomes serve as versatile therapeutic tools, functioning as drug delivery vehicles or facilitating personalized medicine through exosome engineering.
Research has also suggested that exosomal protein cargos can serve as promising biomarkers for predicting OS prognosis. In their study involving 146 patients with OS, Wang et al. identified a significant correlation between the expression of plasma exosomal sentrin SUMO-specific protease 1 (SENP1) and factors such as tumor size, tumor location, necrosis rate, pulmonary metastasis, and surgical stage. Additionally, patients with higher levels of SENP1 expression experienced less favorable outcomes in both overall survival and disease-free survival (DFS) than patients with lower SENP1 expression [56]. Using surface-enhanced Raman scattering (SERS) and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) allows researchers discerning of distinct plasma exosome profiles between OS patients and healthy individuals. Additionally, the MALDI-TOF MS analysis of plasma exosomes from OS patients further identified that seven exosomal protein cargos (IGLV2-23, IGLV4-3, IGLV1-51, IGKV3-15, IGHV4-4, IGLV4-60, and HBA1) are associated with lung metastasis [57][58]. It was observed that the expression of serum exosomal TGFβ was upregulated in OS patients compared to health controls [59]. Likewise, another study demonstrated that the expression level of plasma exosomal PD-L1 and N-cadherin is significantly upregulated in OS patients with lung metastasis, in contrast to those without metastasis. This suggests the possibility of these biomarkers as potential candidates for predicting pulmonary metastasis and tumor progression in OS [60].
In addition to utilizing exosomal RNA and protein cargos as biomarker tools, recent research has begun to explore the potential of exosomal DNA cargos as valuable cancer biomarkers. Thakur and colleagues demonstrated that double-stranded DNA in exosomes derived from tumor cells carries the mutational status of the parent cells, highlighting its potential as a surrogate for detecting genetic mutations in cancer patients [61]. A recent study by Cambier et al. revealed elevated expression of repetitive element DNA, as opposed to RNA levels, within serum exosomes from OS patients compared to the serum exosomes from control subjects. This included the upregulation of HSATI, HSATII, LINE1-P1, and Charlie 3 [62]. Liquid biopsies have emerged as a noninvasive and powerful tool for monitoring tumor progression, predicting prognosis, assessing metastasis, and evaluating drug response in cancer. Considering the remarkable stability of exosomes in various body fluids, the utilization of exosomes as biomarkers holds significant promise for OS.
5. Exosomes as a Vehicle for Drug Delivery in Osteosarcoma Therapy
Considering that chemotherapy remains the primary treatment approach for OS, nanomedicine research harnesses the enhanced permeability and retention (EPR) effect to facilitate drug delivery to OS, thereby decreasing chemotherapy dose, reducing associated toxicity, and targeting specific tumor sites. Apart from the traditional nanocarriers such as nanoparticles, liposomes, and polymeric micelles used for tumor targeting through the EPR effect, emerging carriers employing naturally secreted cellular vesicles exhibit enhanced biocompatibility, reduced immunogenicity, and the ability to cross various biological barriers [63][64][65][66]. Moreover, given exosome capability to transfer cargos that regulates tumorigenesis, angiogenesis, metastasis, and tumor progression, there is a growing research focus on utilizing exosomes as drug delivery vehicles. Exosomes can be engineered to carry bioactive cargos or chemotherapeutic agents (Figure 1). For example, a study by Shimbo et al. reported that upon transfection synthetic miR-143 into BMSCs, miRNA could be encapsulated into exosomes and transferred to OS cells. This exosomal cargo transport was found to inhibit cell migration in OS cells [67]. One of the challenges in utilizing miRNA therapeutics for cancer treatment is the instability of miRNAs and their short systematic half-life, attributed to their rapid renal execration. MiRNAs’ negative charge and hydrophilic characteristics present additional obstacles to crossing the cell membrane [68]. Harnessing exosomes to package miRNAs and facilitate the intercellular transport of the therapeutic cargo presents a potential for enhancing drug delivery outcomes [67]. Wei et al. developed a nanodrug, Exo-Dox, which utilizes exosomes derived from MSCs as nanocarriers for doxorubicin. Their study showed not only significantly improved cellular uptake efficiency but also better anti-tumor effects in OS cells. Notably, their study unveiled that the half-maximal inhibitory concentration (IC50) of Exo-Dox in MG63 cells was found to be lower than free Dox, suggesting the Exo-Dox holds the potential to outperform Dox in effectively treating OS cells [69]. Likewise, the utilization of exosomes derived from bone marrow MSCs to encapsulate doxorubicin exerted heightened tumor suppression and fewer side effects compared to the administration of doxorubicin as a standalone treatment in a xenograft OS model [70]. The authors hypothesized that homing capability of MSC-derived exosomes underlies the findings of both studies. Accordingly, a biodistribution analysis of labeled exosomes derived from human umbilical cord MSCs (HUC-MSCs) in OS tumor-bearing mice revealed that HUC-MSCs exosomes continuously accumulated in OS tumor sites after 24–48 h post-intravenous infusion compared to synthetic nanoparticles. Moreover, dose-dependent inhibition of OS cell proliferation was observed upon exposure to HUC-MSCs exosomes [71]. MSC-derived exosomes are recognized as multifaceted players in OS, with demonstrated roles in mediating OS progression through remodeling the TME remodeling, and functionating as drug carriers to transport therapeutic agents to the tumor site. The impact of MSC-derived exosomes on tumor behavior, whether promoting or inhibiting, is determined by the diverse cargo content present within these exosomes [13][69][70][71][72][73]. In another study, the synthetic agonist of cannabinoid receptors known as WIN was shown to induce a significant exosome secretion, and these exosomes derived from WIN-treated cells were observed to attenuate the migration of OS cells. It is worth noting that the study did not present data on exosomes from untreated cultures [74]. A recent study showed that the surface of exosomes can be engineered for targeted drug delivery. Huang et al. utilized cyclic RGD peptide (cRGD) to modify exosomes, enhancing their tumor-targeting ability. They loaded these modified exosomes with lncRNA-MEG3, a long non-coding RNA with anti-tumor properties. Their findings demonstrated that these engineered exosomes (cRGD-Exo-MEG3) exhibited improved efficiency in targeting OS cells, enhancing anti-tumor effects in vitro and in vivo [75]. Although significant progress has been made in using exosomes as drug delivery systems for cancer treatment, it is crucial to note that these studies are still in the preclinical stage. Numerous aspects require attention and continued investigations, including exosome purification, large-scale good manufacturing practice (GMP) production, and a comprehensive understanding of their mechanisms of action. One key consideration of utilizing exosomes as therapeutics is precisely controlling exosomal cargo content, as various miRNAs, proteins, DNAs, or RNAs might elicit various biological responses. Another factor is the examination of the biodistribution of exosomes to specific organs or tissues. Although early investigations suggest a favorable safety profile, evaluating exosome interaction with immune systems, potential off-target effects, and systematic responses is pivotal. Furthermore, the long-term safety of exosomes as therapeutics needs to be rigorously evaluated to ensure their efficacy and safety profiles throughout the course of cancer treatment. Nevertheless, using exosomes to deliver protein, nucleic acids, or therapeutics remains promising for advancing drug development in OS. As the field advances, thorough investigations into these aspects will not only gain deeper insights into the therapeutic potential of exosomes, but also guide us to establish robust and effective clinical applications.
References
- Jerez, S.; Araya, H.; Hevia, D.; Irarrázaval, C.E.; Thaler, R.; van Wijnen, A.J.; Galindo, M. Extracellular Vesicles from Osteosarcoma Cell Lines Contain miRNAs Associated with Cell Adhesion and Apoptosis. Gene 2019, 710, 246–257.
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR Addiction in an in Vivo Model of microRNA-21-Induced Pre-B-Cell Lymphoma. Nature 2010, 467, 86–90.
- Si, M.-L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.-Y. miR-21-Mediated Tumor Growth. Oncogene 2007, 26, 2799–2803.
- Chan, J.A.; Krichevsky, A.M.; Kosik, K.S. MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res. 2005, 65, 6029–6033.
- Raimondi, L.; De Luca, A.; Gallo, A.; Costa, V.; Russelli, G.; Cuscino, N.; Manno, M.; Raccosta, S.; Carina, V.; Bellavia, D.; et al. Osteosarcoma Cell-Derived Exosomes Affect Tumor Microenvironment by Specific Packaging of microRNAs. Carcinogenesis 2020, 41, 666–677.
- Qi, J.; Zhang, R.; Wang, Y. Exosomal miR-21-5p Derived from Bone Marrow Mesenchymal Stem Cells Promote Osteosarcoma Cell Proliferation and Invasion by Targeting PIK3R1. J. Cell. Mol. Med. 2021, 25, 11016–11030.
- Li, C.; Xu, B.; Miu, X.; Deng, Z.; Liao, H.; Hao, L. Inhibition of miRNA-21 Attenuates the Proliferation and Metastasis of Human Osteosarcoma by Upregulating PTEN. Exp. Ther. Med. 2018, 15, 1036–1040.
- Hu, X.; Li, L.; Lu, Y.; Yu, X.; Chen, H.; Yin, Q.; Zhang, Y. miRNA-21 Inhibition Inhibits Osteosarcoma Cell Proliferation by Targeting PTEN and Regulating the TGF-Β1 Signaling Pathway. Oncol. Lett. 2018, 16, 4337–4342.
- Han, F.; Pu, P.; Wang, C.; Ding, X.; Zhu, Z.; Xiang, W.; Wang, W. Osteosarcoma Cell-Derived Exosomal miR-1307 Promotes Tumorgenesis via Targeting AGAP1. Biomed. Res. Int. 2021, 2021, 7358153.
- Ye, Z.; Zheng, Z.; Peng, L. MicroRNA Profiling of Serum Exosomes in Patients with Osteosarcoma by High-Throughput Sequencing. J. Investig. Med. 2020, 68, 893–901.
- Gong, L.; Bao, Q.; Hu, C.; Wang, J.; Zhou, Q.; Wei, L.; Tong, L.; Zhang, W.; Shen, Y. Exosomal miR-675 from Metastatic Osteosarcoma Promotes Cell Migration and Invasion by Targeting CALN1. Biochem. Biophys. Res. Commun. 2018, 500, 170–176.
- Wang, J.-W.; Wu, X.-F.; Gu, X.-J.; Jiang, X.-H. Exosomal miR-1228 from Cancer-Associated Fibroblasts Promotes Cell Migration and Invasion of Osteosarcoma by Directly Targeting SCAI. Oncol. Res. 2019, 27, 979–986.
- Qin, F.; Tang, H.; Zhang, Y.; Zhang, Z.; Huang, P.; Zhu, J. Bone Marrow-Derived Mesenchymal Stem Cell-Derived Exosomal microRNA-208a Promotes Osteosarcoma Cell Proliferation, Migration, and Invasion. J. Cell. Physiol. 2020, 235, 4734–4745.
- Liu, W.; Wang, B.; Duan, A.; Shen, K.; Zhang, Q.; Tang, X.; Wei, Y.; Tang, J.; Zhang, S. Exosomal Transfer of miR-769-5p Promotes Osteosarcoma Proliferation and Metastasis by Targeting DUSP16. Cancer Cell Int. 2021, 21, 541.
- Yoshida, A.; Fujiwara, T.; Uotani, K.; Morita, T.; Kiyono, M.; Yokoo, S.; Hasei, J.; Nakata, E.; Kunisada, T.; Ozaki, T. Clinical and Functional Significance of Intracellular and Extracellular microRNA-25-3p in Osteosarcoma. Acta Med. Okayama 2018, 72, 165–174.
- Zhang, P.; Zhang, J.; Quan, H.; Wang, J.; Liang, Y. MicroRNA-143 Expression Inhibits the Growth and the Invasion of Osteosarcoma. J. Orthop. Surg. Res. 2022, 17, 236.
- Dong, X.; Lv, B.; Li, Y.; Cheng, Q.; Su, C.; Yin, G. MiR-143 Regulates the Proliferation and Migration of Osteosarcoma Cells through Targeting MAPK7. Arch. Biochem. Biophys. 2017, 630, 47–53.
- Li, W.; Wu, H.; Li, Y.; Pan, H.; Meng, T.; Wang, X. MicroRNA-143 Promotes Apoptosis of Osteosarcoma Cells by Caspase-3 Activation via Targeting Bcl-2. Biomed. Pharmacother. 2016, 80, 8–15.
- Osaki, M.; Takeshita, F.; Sugimoto, Y.; Kosaka, N.; Yamamoto, Y.; Yoshioka, Y.; Kobayashi, E.; Yamada, T.; Kawai, A.; Inoue, T.; et al. MicroRNA-143 Regulates Human Osteosarcoma Metastasis by Regulating Matrix Metalloprotease-13 Expression. Mol. Ther. 2011, 19, 1123–1130.
- Holenstein, C.N.; Horvath, A.; Schär, B.; Schoenenberger, A.D.; Bollhalder, M.; Goedecke, N.; Bartalena, G.; Otto, O.; Herbig, M.; Guck, J.; et al. The Relationship between Metastatic Potential and in Vitro Mechanical Properties of Osteosarcoma Cells. MBoC 2019, 30, 887–898.
- Troyer, R.M.; Ruby, C.E.; Goodall, C.P.; Yang, L.; Maier, C.S.; Albarqi, H.A.; Brady, J.V.; Bathke, K.; Taratula, O.; Mourich, D.; et al. Exosomes from Osteosarcoma and Normal Osteoblast Differ in Proteomic Cargo and Immunomodulatory Effects on T Cells. Exp. Cell Res. 2017, 358, 369–376.
- Deng, C.; Xu, Y.; Fu, J.; Zhu, X.; Chen, H.; Xu, H.; Wang, G.; Song, Y.; Song, G.; Lu, J.; et al. Reprograming the Tumor Immunologic Microenvironment Using Neoadjuvant Chemotherapy in Osteosarcoma. Cancer Sci. 2020, 111, 1899–1909.
- Dumars, C.; Ngyuen, J.-M.; Gaultier, A.; Lanel, R.; Corradini, N.; Gouin, F.; Heymann, D.; Heymann, M.-F. Dysregulation of Macrophage Polarization Is Associated with the Metastatic Process in Osteosarcoma. Oncotarget 2016, 7, 78343–78354.
- Qian, B.-Z.; Pollard, J.W. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell 2010, 141, 39–51.
- Mosser, D.M.; Edwards, J.P. Exploring the Full Spectrum of Macrophage Activation. Nat. Rev. Immunol. 2008, 8, 958–969.
- Wolf-Dennen, K.; Gordon, N.; Kleinerman, E.S. Exosomal Communication by Metastatic Osteosarcoma Cells Modulates Alveolar Macrophages to an M2 Tumor-Promoting Phenotype and Inhibits Tumoricidal Functions. OncoImmunology 2020, 9, 1747677.
- Cheng, Z.; Wang, L.; Wu, C.; Huang, L.; Ruan, Y.; Xue, W. Tumor-Derived Exosomes Induced M2 Macrophage Polarization and Promoted the Metastasis of Osteosarcoma Cells Through Tim-3. Arch. Med. Res. 2021, 52, 200–210.
- Wang, B.; Wang, X.; Li, P.; Niu, X.; Liang, X.; Liu, G.; Liu, Z.; Ge, H. Osteosarcoma Cell-Derived Exosomal ELFN1-AS1 Mediates Macrophage M2 Polarization via Sponging miR-138-5p and miR-1291 to Promote the Tumorgenesis of Osteosarcoma. Front. Oncol. 2022, 12, 881022.
- Zhong, L.; Liao, D.; Li, J.; Liu, W.; Wang, J.; Zeng, C.; Wang, X.; Cao, Z.; Zhang, R.; Li, M.; et al. Rab22a-NeoF1 Fusion Protein Promotes Osteosarcoma Lung Metastasis through Its Secretion into Exosomes. Signal Transduct. Target. Ther. 2021, 6, 59.
- Liu, W.; Long, Q.; Zhang, W.; Zeng, D.; Hu, B.; Liu, S.; Chen, L. miRNA-221-3p Derived from M2-Polarized Tumor-Associated Macrophage Exosomes Aggravates the Growth and Metastasis of Osteosarcoma through SOCS3/JAK2/STAT3 Axis. Aging 2021, 13, 19760–19775.
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 Contributes to Immunosuppression and Is Associated with Anti-PD-1 Response. Nature 2018, 560, 382–386.
- Kim, J.W.; Wieckowski, E.; Taylor, D.D.; Reichert, T.E.; Watkins, S.; Whiteside, T.L. Fas Ligand–Positive Membranous Vesicles Isolated from Sera of Patients with Oral Cancer Induce Apoptosis of Activated T Lymphocytes. Clin. Cancer Res. 2005, 11, 1010–1020.
- Zhang, L.; Xue, L.; Wu, Y.; Wu, Q.; Ren, H.; Song, X. Exosomes Loaded with Programmed Death Ligand-1 Promote Tumor Growth by Immunosuppression in Osteosarcoma. Bioengineered 2021, 12, 9520–9530.
- Dorayappan, K.D.P.; Wanner, R.; Wallbillich, J.J.; Saini, U.; Zingarelli, R.; Suarez, A.A.; Cohn, D.E.; Selvendiran, K. Hypoxia-Induced Exosomes Contribute to a More Aggressive and Chemoresistant Ovarian Cancer Phenotype: A Novel Mechanism Linking STAT3/Rab Proteins. Oncogene 2018, 37, 3806–3821.
- Lobb, R.J.; van Amerongen, R.; Wiegmans, A.; Ham, S.; Larsen, J.E.; Möller, A. Exosomes Derived from Mesenchymal Non-Small Cell Lung Cancer Cells Promote Chemoresistance. Int. J. Cancer 2017, 141, 614–620.
- Hu, Y.; Yan, C.; Mu, L.; Huang, K.; Li, X.; Tao, D.; Wu, Y.; Qin, J. Fibroblast-Derived Exosomes Contribute to Chemoresistance through Priming Cancer Stem Cells in Colorectal Cancer. PLoS ONE 2015, 10, e0125625.
- Ji, R.; Zhang, B.; Zhang, X.; Xue, J.; Yuan, X.; Yan, Y.; Wang, M.; Zhu, W.; Qian, H.; Xu, W. Exosomes Derived from Human Mesenchymal Stem Cells Confer Drug Resistance in Gastric Cancer. Cell Cycle 2015, 14, 2473–2483.
- Shao, H.; Chung, J.; Lee, K.; Balaj, L.; Min, C.; Carter, B.S.; Hochberg, F.H.; Breakefield, X.O.; Lee, H.; Weissleder, R. Chip-Based Analysis of Exosomal mRNA Mediating Drug Resistance in Glioblastoma. Nat. Commun. 2015, 6, 6999.
- Chen, W.; Liu, X.; Lv, M.; Chen, L.; Zhao, J.; Zhong, S.; Ji, M.; Hu, Q.; Luo, Z.; Wu, J.; et al. Exosomes from Drug-Resistant Breast Cancer Cells Transmit Chemoresistance by a Horizontal Transfer of microRNAs. PLoS ONE 2014, 9, e95240.
- Torreggiani, E.; Roncuzzi, L.; Perut, F.; Zini, N.; Baldini, N. Multimodal Transfer of MDR by Exosomes in Human Osteosarcoma. Int. J. Oncol. 2016, 49, 189–196.
- Hu, X.; Wen, Y.; Tan, L.; Wang, J.; Tang, F.; Wang, Y.; Zheng, C.; Zhang, Y.; Gong, T.; Min, L. Exosomal Long Non-Coding RNA ANCR Mediates Drug Resistance in Osteosarcoma. Front. Oncol. 2022, 11, 735254.
- Pan, Y.; Lin, Y.; Mi, C. Cisplatin-Resistant Osteosarcoma Cell-Derived Exosomes Confer Cisplatin Resistance to Recipient Cells in an Exosomal Circ_103801-Dependent Manner. Cell Biol. Int. 2021, 45, 858–868.
- Zhan, H.; Xiao, J.; Wang, P.; Mo, F.; Li, K.; Guo, F.; Yu, X.; Liu, X.; Zhang, B.; Dai, M.; et al. Exosomal CTCF Confers Cisplatin Resistance in Osteosarcoma by Promoting Autophagy via the IGF2-AS/miR-579-3p/MSH6 Axis. J. Oncol. 2022, 2022, e9390611.
- Yati, S.; Silathapanasakul, A.; Thakaeng, C.; Chanasakulniyom, M.; Songtawee, N.; Porntadavity, S.; Pothacharoen, P.; Pruksakorn, D.; Kongtawelert, P.; Yenchitsomanus, P.; et al. Extracellular Vesicle-Mediated IL-1 Signaling in Response to Doxorubicin Activates PD-L1 Expression in Osteosarcoma Models. Cells 2022, 11, 1042.
- Liu, J.; Quan, L.; Zhang, C.; Liu, A.; Tong, D.; Wang, J. Over-Activated PD-1/PD-L1 Axis Facilitates the Chemoresistance of Diffuse Large B-Cell Lymphoma Cells to the CHOP Regimen. Oncol. Lett. 2018, 15, 3321–3328.
- Black, M.; Barsoum, I.B.; Truesdell, P.; Cotechini, T.; Macdonald-Goodfellow, S.K.; Petroff, M.; Siemens, D.R.; Koti, M.; Craig, A.W.B.; Graham, C.H. Activation of the PD-1/PD-L1 Immune Checkpoint Confers Tumor Cell Chemoresistance Associated with Increased Metastasis. Oncotarget 2016, 7, 10557–10567.
- Yan, F.; Pang, J.; Peng, Y.; Molina, J.R.; Yang, P.; Liu, S. Elevated Cellular PD1/PD-L1 Expression Confers Acquired Resistance to Cisplatin in Small Cell Lung Cancer Cells. PLoS ONE 2016, 11, e0162925.
- Bao, Q.; Gong, L.; Wang, J.; Wen, J.; Shen, Y.; Zhang, W. Extracellular Vesicle RNA Sequencing Reveals Dramatic Transcriptomic Alterations Between Metastatic and Primary Osteosarcoma in a Liquid Biopsy Approach. Ann. Surg. Oncol. 2018, 25, 2642–2651.
- Fujiwara, T.; Uotani, K.; Yoshida, A.; Morita, T.; Nezu, Y.; Kobayashi, E.; Yoshida, A.; Uehara, T.; Omori, T.; Sugiu, K.; et al. Clinical Significance of Circulating miR-25-3p as a Novel Diagnostic and Prognostic Biomarker in Osteosarcoma. Oncotarget 2017, 8, 33375–33392.
- Zhang, K.; Dong, C.; Chen, M.; Yang, T.; Wang, X.; Gao, Y.; Wang, L.; Wen, Y.; Chen, G.; Wang, X.; et al. Extracellular Vesicle-Mediated Delivery of miR-101 Inhibits Lung Metastasis in Osteosarcoma. Theranostics 2020, 10, 411–425.
- Xu, J.-F.; Wang, Y.-P.; Zhang, S.-J.; Chen, Y.; Gu, H.-F.; Dou, X.-F.; Xia, B.; Bi, Q.; Fan, S.-W. Exosomes Containing Differential Expression of microRNA and mRNA in Osteosarcoma That Can Predict Response to Chemotherapy. Oncotarget 2017, 8, 75968–75978.
- Cuscino, N.; Raimondi, L.; De Luca, A.; Carcione, C.; Russelli, G.; Conti, L.; Baldi, J.; Conaldi, P.G.; Giavaresi, G.; Gallo, A. Gathering Novel Circulating Exosomal microRNA in Osteosarcoma Cell Lines and Possible Implications for the Disease. Cancers 2019, 11, 1924.
- Zhang, H.; Wang, J.; Ren, T.; Huang, Y.; Yu, Y.; Chen, C.; Huang, Q.; Guo, W. LncRNA CASC15 Is Upregulated in Osteosarcoma Plasma Exosomes and CASC15 Knockdown Inhibits Osteosarcoma Progression by Regulating miR-338-3p/RAB14 Axis. Onco Targets Ther. 2020, 13, 12055–12066.
- Li, S.; Pei, Y.; Wang, W.; Liu, F.; Zheng, K.; Zhang, X. Extracellular Nanovesicles-transmitted Circular RNA Has_circ_0000190 Suppresses Osteosarcoma Progression. J. Cell. Mol. Med. 2020, 24, 2202–2214.
- Huo, S.; Dou, D. Circ_0056285 Regulates Proliferation, Apoptosis and Glycolysis of Osteosarcoma Cells via miR-1244/TRIM44 Axis. Cancer Manag. Res. 2021, 13, 1257–1270.
- Wang, L.; Wu, J.; Song, S.; Chen, H.; Hu, Y.; Xu, B.; Liu, J. Plasma Exosome-Derived Sentrin SUMO-Specific Protease 1: A Prognostic Biomarker in Patients with Osteosarcoma. Front. Oncol. 2021, 11, 625109.
- Han, Z.; Peng, C.; Yi, J.; Wang, Y.; Liu, Q.; Yang, Y.; Long, S.; Qiao, L.; Shen, Y. Matrix-Assisted Laser Desorption Ionization Mass Spectrometry Profiling of Plasma Exosomes Evaluates Osteosarcoma Metastasis. iScience 2021, 24, 102906.
- Han, Z.; Yi, J.; Yang, Y.; Li, D.; Peng, C.; Long, S.; Peng, X.; Shen, Y.; Liu, B.; Qiao, L. SERS and MALDI-TOF MS Based Plasma Exosome Profiling for Rapid Detection of Osteosarcoma. Analyst 2021, 146, 6496–6505.
- Baglio, S.R.; Lagerweij, T.; Pérez-Lanzón, M.; Ho, X.D.; Léveillé, N.; Melo, S.A.; Cleton-Jansen, A.-M.; Jordanova, E.S.; Roncuzzi, L.; Greco, M.; et al. Blocking Tumor-Educated MSC Paracrine Activity Halts Osteosarcoma Progression. Clin. Cancer Res. 2017, 23, 3721–3733.
- Wang, J.; Zhang, H.; Sun, X.; Wang, X.; Ren, T.; Huang, Y.; Zhang, R.; Zheng, B.; Guo, W. Exosomal PD-L1 and N-Cadherin Predict Pulmonary Metastasis Progression for Osteosarcoma Patients. J. Nanobiotechnol. 2020, 18, 151.
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J.; et al. Double-Stranded DNA in Exosomes: A Novel Biomarker in Cancer Detection. Cell Res. 2014, 24, 766–769.
- Cambier, L.; Stachelek, K.; Triska, M.; Jubran, R.; Huang, M.; Li, W.; Zhang, J.; Li, J.; Cobrinik, D. Extracellular Vesicle-Associated Repetitive Element DNAs as Candidate Osteosarcoma Biomarkers. Sci. Rep. 2021, 11, 94.
- Elliott, R.O.; He, M. Unlocking the Power of Exosomes for Crossing Biological Barriers in Drug Delivery. Pharmaceutics 2021, 13, 122.
- Yong, T.; Zhang, X.; Bie, N.; Zhang, H.; Zhang, X.; Li, F.; Hakeem, A.; Hu, J.; Gan, L.; Santos, H.A.; et al. Tumor Exosome-Based Nanoparticles Are Efficient Drug Carriers for Chemotherapy. Nat. Commun. 2019, 10, 3838.
- Zhu, X.; Badawi, M.; Pomeroy, S.; Sutaria, D.S.; Xie, Z.; Baek, A.; Jiang, J.; Elgamal, O.A.; Mo, X.; Perle, K.L.; et al. Comprehensive Toxicity and Immunogenicity Studies Reveal Minimal Effects in Mice Following Sustained Dosing of Extracellular Vesicles Derived from HEK293T Cells. J. Extracell. Vesicles 2017, 6, 1324730.
- Chen, C.C.; Liu, L.; Ma, F.; Wong, C.W.; Guo, X.E.; Chacko, J.V.; Farhoodi, H.P.; Zhang, S.X.; Zimak, J.; Ségaliny, A.; et al. Elucidation of Exosome Migration Across the Blood–Brain Barrier Model In Vitro. Cell. Mol. Bioeng. 2016, 9, 509–529.
- Shimbo, K.; Miyaki, S.; Ishitobi, H.; Kato, Y.; Kubo, T.; Shimose, S.; Ochi, M. Exosome-Formed Synthetic microRNA-143 Is Transferred to Osteosarcoma Cells and Inhibits Their Migration. Biochem. Biophys. Res. Commun. 2014, 445, 381–387.
- Segal, M.; Biscans, A.; Gilles, M.-E.; Anastasiadou, E.; Luca, R.D.; Lim, J.; Khvorova, A.; Slack, F.J. Hydrophobically Modified Let-7b miRNA Enhances Biodistribution to NSCLC and Downregulates HMGA2 In Vivo. Mol. Ther. Nucleic Acids 2020, 19, 267–277.
- Wei, H.; Chen, J.; Wang, S.; Fu, F.; Zhu, X.; Wu, C.; Liu, Z.; Zhong, G.; Lin, J. A Nanodrug Consisting of Doxorubicin and Exosome Derived from Mesenchymal Stem Cells for Osteosarcoma Treatment In Vitro. Int. J. Nanomed. 2019, 14, 8603–8610.
- Wang, J.; Li, M.; Jin, L.; Guo, P.; Zhang, Z.; Zhanghuang, C.; Tan, X.; Mi, T.; Liu, J.; Wu, X.; et al. Exosome Mimetics Derived from Bone Marrow Mesenchymal Stem Cells Deliver Doxorubicin to Osteosarcoma in Vitro and in Vivo. Drug Deliv. 2022, 29, 3291–3303.
- Abello, J.; Nguyen, T.D.T.; Marasini, R.; Aryal, S.; Weiss, M.L. Biodistribution of Gadolinium- and near Infrared-Labeled Human Umbilical Cord Mesenchymal Stromal Cell-Derived Exosomes in Tumor Bearing Mice. Theranostics 2019, 9, 2325–2345.
- Zhang, H.; Wang, J.; Ren, T.; Huang, Y.; Liang, X.; Yu, Y.; Wang, W.; Niu, J.; Guo, W. Bone Marrow Mesenchymal Stem Cell-Derived Exosomal miR-206 Inhibits Osteosarcoma Progression by Targeting TRA2B. Cancer Lett. 2020, 490, 54–65.
- Li, F.; Chen, X.; Shang, C.; Ying, Q.; Zhou, X.; Zhu, R.; Lu, H.; Hao, X.; Dong, Q.; Jiang, Z. Bone Marrow Mesenchymal Stem Cells-Derived Extracellular Vesicles Promote Proliferation, Invasion and Migration of Osteosarcoma Cells via the lncRNA MALAT1/miR-143/NRSN2/Wnt/β-Catenin Axis. Onco Targets Ther. 2021, 14, 737–749.
- Notaro, A.; Emanuele, S.; Geraci, F.; D’Anneo, A.; Lauricella, M.; Calvaruso, G.; Giuliano, M. WIN55,212-2-Induced Expression of Mir-29b1 Favours the Suppression of Osteosarcoma Cell Migration in a SPARC-Independent Manner. Int. J. Mol. Sci. 2019, 20, 5235.
- Huang, X.; Wu, W.; Jing, D.; Yang, L.; Guo, H.; Wang, L.; Zhang, W.; Pu, F.; Shao, Z. Engineered Exosome as Targeted lncRNA MEG3 Delivery Vehicles for Osteosarcoma Therapy. J. Control. Release 2022, 343, 107–117.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
549
Revisions:
2 times
(View History)
Update Date:
15 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




