Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Małgorzata Wrzosek | -- | 1712 | 2024-01-08 17:54:52 | | | |
| 2 | Wendy Huang | Meta information modification | 1712 | 2024-01-09 12:41:40 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zarembska, E.; Ślusarczyk, K.; Wrzosek, M. Methylenetetrahydrofolate Reductase Polymorphism and Regulation of Its Activity. Encyclopedia. Available online: https://encyclopedia.pub/entry/53568 (accessed on 08 February 2026).
Zarembska E, Ślusarczyk K, Wrzosek M. Methylenetetrahydrofolate Reductase Polymorphism and Regulation of Its Activity. Encyclopedia. Available at: https://encyclopedia.pub/entry/53568. Accessed February 08, 2026.
Zarembska, Emilia, Klaudia Ślusarczyk, Małgorzata Wrzosek. "Methylenetetrahydrofolate Reductase Polymorphism and Regulation of Its Activity" Encyclopedia, https://encyclopedia.pub/entry/53568 (accessed February 08, 2026).
Zarembska, E., Ślusarczyk, K., & Wrzosek, M. (2024, January 08). Methylenetetrahydrofolate Reductase Polymorphism and Regulation of Its Activity. In Encyclopedia. https://encyclopedia.pub/entry/53568
Zarembska, Emilia, et al. "Methylenetetrahydrofolate Reductase Polymorphism and Regulation of Its Activity." Encyclopedia. Web. 08 January, 2024.
Copy Citation
Methylenetetrahydrofolate reductase (MTHFR) is an enzyme encoded by the MTHFR gene composed of 12 exons and located on chromosome 1p36.22. Its total length is 20,374 bp. Methylenetetrahydrofolate reductase (MTHFR) is a key regulatory enzyme in the one-carbon cycle. This enzyme is essential for the metabolism of methionine, folate, and RNA, as well as for the production of proteins, DNA, and RNA. MTHFR catalyses the irreversible conversion of 5,10-methylenetetrahydrofolate to its active form, 5-methyltetrahydrofolate, a co-substrate for homocysteine remethylation to methionine.
MTHFR
folate
polymorphism
activity
protein
1. Methylenetetrahydrofolate Reductase (MTHFR)
Methylenetetrahydrofolate reductase (MTHFR) is an enzyme encoded by the MTHFR gene composed of 12 exons and located on chromosome 1p36.22. Its total length is 20,374 bp [1]. Characteristic elements, such as SP1, AP1, AP2, CAAT, or GC, are involved in the regulation of the expression of this gene, but there are no TATA-box elements. The structure of this promoter region is shared by other genes involved in homocysteine (Hcy) metabolism, including cystathionine β-synthase (CBS), methionine synthase (MS), and methionine synthase reductase coding genes [2].
During MTHFR transcription, alternative splicing occurs, resulting in three different mRNA molecules of 7074, 7018, and 7071 bp, which encode polypeptides of 697, 656, and 696 amino acids, respectively [1]. On this basis, three variants of the human MTHFR transcript have been distinguished: MTHFR 1, 2, and 3, which differ at the end of 5′ [3]. As mentioned above, the presence of the transcript’s different sizes was due to alternative transcription start sites and the use of polyadenylation signal sequences [4]. Western blot analysis discovered a major polypeptide of approximately 77 kDa in human tissues, while a protein isoform of approximately 70 kDa was detected only in human liver tissue [5][6].
MTHFR is an FAD-dependent enzyme that plays a significant role in the metabolism of folate and Hcy, both of which are based on folic acid and other vitamins in the B group. This enzyme catalyses the NADPH-linked reduction of 5,10-methylenetetrahydrofolate (5,10-methylene-THF) to 5-methyltetrahydrofolate (5-methyl-THF). The last molecule is a methyl group donor for the conversion of Hcy to methionine (Met) in the reaction catalysed by methionine synthase (MS) (Figure 1). Vitamin B12 acts as a cofactor during this process. Met is then converted to S-adenosylmethionine (SAM), which is a crucial methyl group donor for various reactions in the body, including the methylation of DNA [7], RNA, histones, phospholipids, choline, sphingomyelin, acetylcholine, and other neurotransmitters [8]. Protein carboxymethylation may be involved in the repair of ageing proteins; also, heat shock proteins are methylated in response to stress [9]. In addition, Hcy can be converted to cysteine through a trans-sulphuration process involving the enzyme CBS and vitamin B6. Under Met deficiency conditions, CBS is not activated and MTHFR is not inhibited by SAM. As a result, Hcy is converted back into Met, while cysteine contributes to glutathione synthesis or is degraded to taurine (Figure 1) [10].
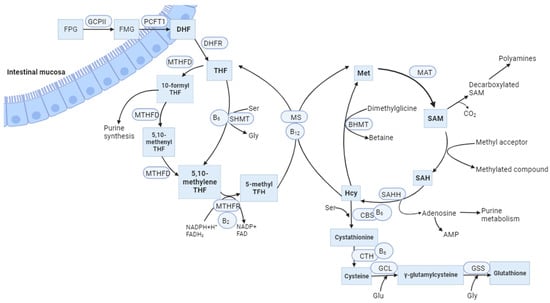
Figure 1. The balance between the folate cycle and the methionine cycle is affected by vitamin B12. FPG, Folate polyglutamates; FMG, Folate monoglutamates; GCPII, glutamate carboxypeptidase II; PCFT1, proton-coupled folate transporter; DHF, dihydrofolate; DHRF, dihydrofolate reductase; THF, tetrahydrofolate; MTHFD, methylenetetrahydrofolate dehydrogenase; SHMT, serinehydroxymethyl transferase; Ser, Serine; Gly, Glycine; MTHFR, methylenetetrahydrofolate reductase; NADPH, nicotinamide adenine dinucleotide phosphate; FADH2, dihydroflavine-adeninedinucleotide; MS, methionine synthase; Met, Methionine; MAT, methionine adenosyltransferase; SAM, S-adenosylmethionine; SAH, S-adenosylhomocysteine; SAHH, S-adenosylhomocysteine hydrolase; BHMT, Betaine-homocysteine methyltransferase; CBS, Cystathionine-β-synthase; CTH, Cystathionine gamma lyase; GCL, glutamate cysteine ligase; GSS, Glutathione synthetase; Glu, Glutamic acid; B2, vitamin B2 (riboflavin); B6, vitamin B6 (pyridoxine); B12, vitamin B12 (cobalamin).
2. MTHFR Polymorphism
The first documentation of MTHFR involvement in disease came from the research conducted by Mudd et al. in 1972 [11]. They identified patients with homocystinuria, which was attributed to a significant deficiency in MTHFR obtained from fibroblasts. Kang et al. in 1988 described decreased activity and increased thermolability of the MTHFR enzyme in lymphocyte extracts obtained from patients with ischemic heart disease (IHD) [12]. Some of them had a decrease in enzyme activity by up to 75% and an increase in total Hcy levels, but this was not the case in patients with high levels of folate and vitamin B12 [13]. The variations observed between individuals suggest the existence of genetic diversity within MTHFR. Currently, several dozen polymorphisms have been identified, with the most studied being the C677T (rs1801133) and the A1298C polymorphism (rs180113) [14][15]. The first one involves a substitution of cytosine to thymine at position 677 within exon 4, leading to a substitution of alanine to valine at position 222 within the catalytic domain of the MTHFR. This site is crucial in terms of the binding of flavin adenine dinucleotides (FAD) and enzyme stability. The 677T allele encodes a thermolabile enzyme with reduced activity and less affinity for its cofactor, FAD. Each copy of the 677T allele results in 35% reduced enzyme activity. The 677TT homozygotes are believed to have reduced levels of active folate (5-methyl-THF) and increased plasma levels of Hcy because it cannot be remethylated to Met [6].
The prevalence of the C677T polymorphism varies by ethnic group and geographic location and has a relatively high frequency worldwide. A meta-analysis of population-based studies revealed that the worldwide prevalence of the T allele was estimated to be 24.0%, while the global occurrence of the TT genotype was 7.7%. However, upon closer examination of different subgroups, it became evident that the prevalence of the T allele exhibited significant variation: 10.3% in Africans, 31.2% in North Americans, 27.8% in South Americans, 19.7% in Asians, 20.5% in Australians, and 34.1% in Europeans. The prevalence of both the T allele and the TT genotype was lowest among Africans and highest among Europeans. The occurrence of the T allele differs significantly between the Asian population. Research carried out within Asian populations showed that this particular gene polymorphism showed a notably higher prevalence in East Asian countries (44.7% in China, 40.3% in the Republic of Korea, and 39.9% in Japan). On the contrary, South Asian countries had a lower prevalence (11.4% in India, 16% in Pakistan, and 4.5% in Sri Lanka) [16].
Another common MTHFR polymorphism is 1298A>C. However, the presence of this variant does not lead to elevated levels of Hcy in heterozygous or homozygous individuals. Instead, the combined heterozygosity of 1298A>C and 677C>T produces similar results to having a TT genotype [17].
3. Regulation of MTHFR Activity
Folate metabolism and the methionine cycle share a common step involving the MTHFR reaction, so regulation of reductase activity is crucial for maintaining reference Met and SAM concentrations in cells. MTHFR activity is inhibited by dihydrofolate (DHF) and its polyglutamate analogues [18]. The human MTHFR protein comprises multiple domains, including an N-terminal catalytic domain, which houses a conserved serine-rich region, and a C-terminal regulatory domain (Figure 2). The catalytic domain is connected to the regulatory domain by a linker sequence (“linker region”). The regulatory domain plays a role in binding to SAM, which subsequently functions as an allosteric inhibitor of reductase activity [19]. The effect of the inhibition reaction is very slow and can be reversed by the attachment of SAH, the demethylated form of SAM [20].
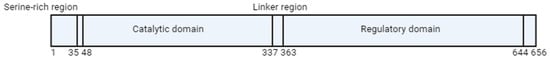
Figure 2. Schematic representation of MTHFR protein. The numbers given represent amino acids in human MTHFR.
The enzyme activity is also affected by multiple phosphorylation of the serine-rich region [21]. One of the phosphorylation sites is Thr 34. The substitution of threonine to alanine at position 34 completely blocks phosphorylation, suggesting that Thr 34 serves as the initiation site for this process. Yamada et al. [20] expressed the mutant Thr34Ala in baculovirus-infected insect cells infected with baculoviruses and compared its enzymatic properties with the wild-type enzyme. MTHFR was treated with alkaline phosphatase, which removes seven phosphoryl groups from the enzyme. The wild-type exposed to alkaline phosphatase and the mutant enzyme had higher activity compared to the wild-type enzyme not treated with phosphatase, and they were also less sensitive to inhibition by SAM [22]. Phosphorylation probably protects SAM from spontaneous degradation to SAH and may induce a conformational change in the MTHFR enzyme. Furthermore, SAM may affect the stability of the linkage to FAD, an essential cofactor of MTHFR. The presence of mutations or polymorphisms in the MTHFR gene that affect the function of FAD may have similar effects [21].
The molecular basis of the enzymatic regulation remained unknown until 2018. The article by Froese et al. identified the aforementioned “linker region”, which links two processes: SAM binding to the regulatory domain and inhibition in the catalytic domain (Figure 2). Moreover, these processes are individually mediated by regions more than 300 amino acids apart [21].
An indicator of the ability of cells to methylate DNA or form compounds requiring methyl groups is the SAM/SAH ratio. Under conditions of low SAM/SAH ratio (methyl donor deficiency), MTHFR is activated, leading to an increase in the concentration of active folate (5-methyl-THF). As a result, the concentration of SAM in the cell increases. A high SAM/SAH ratio leads to efficient and effective methylation, resulting in SAM-mediated allosteric inhibition of the enzyme, which reduces the concentration of 5-methyl-THF and decreases the activity of the methionine cycle, and thus also the production of SAM [21].
Observations of MTHFR regulation by phosphorylation were validated in a study by Zheng et al. [23].They investigated the activity of the DYRK1A/2 and GSK3A/B kinases responsible for multisite phosphorylation of MTHFR and its physiological significance in cells. To confirm that the phosphorylated MTHFR is less active than the non-phosphorylated form, under physiological SAM concentration conditions (1–3 μM), mutant MTHFR knock-in lines were created using the CRISPR method (Clustered Regularly Interspaced Short Palindromic Repeats). The enzyme with a mutation (completely devoid of phosphorylation) was compared to the original parental cell lines. The parental cell lines showed an increase in 5-methyl-THF production in response to Hcy treatment, while the knockin cell lines had high basal levels of 5-methyl-THF and did not respond to Hcy treatment. The results suggest that multisite phosphorylation of the MTHFR enzyme is associated with SAM attachment to inhibit MTHFR activity in cells [23].
In summary, the physiological function of multisite phosphorylation is considered to be as follows: under conditions of high Met concentration, it provides maximum inhibition of MTHFR by SAM, so that one-carbon units are “spared” for key processes, such as purine and deoxythymidine monophosphate (dTMP) synthesis. On the contrary, under low Met concentration conditions, MTHFR is dephosphorylated and becomes more active, leading to a diversion of more one-carbon units into the Met and SAM synthesis pathway. Dephosphorylation and activation of MTHFR may also serve as a cellular response to hyperhomocysteinemia (HHcy), ensuring sufficient concentrations of active folate for the remethylation of Hcy and the elimination of toxic effects caused by this amino acid [23].
References
- National Center for Biotechnology Information (NCBI). 2023. Available online: https://www.ncbi.nlm.nih.gov/ (accessed on 8 October 2023).
- Homberger, A.; Linnebank, M.; Winter, C.; Willenbring, H.; Marquardt, T.; Harms, E.; Koch, H.G. Genomic structure and transcript variants of the human methylenertolate reductase gene. Eur. J. Hum. Genet. 2000, 8, 725–729.
- Goyette, P.; Pai, A.; Milos, R.; Frosst, P.; Tran, P.; Chen, Z.; Chan, M.; Rozen, R. Gene structure of human and mouse methylenetetrahydrofolate reductase (MTHFR). Mamm. Genome 1998, 9, 652–656.
- Gaughan, D.J.; Barbaux, S.; Kluijtmans, L.A.; Whitehead, A.S. The human and mouse methylenetetrahydrofolate reductase (MTHFR) genes: Genomic organization, mRNA structure and linkage to the CLCN6 gene. Gene 2000, 257, 279–289.
- Tran, P.; Leclerc, D.; Chan, M.; Pai, A.; Hiou-Tim, F.; Wu, Q.; Goyette, P.; Artigas, C.; Milos, R.; Rozen, R. Multiple transcription start sites and alternative splicing in the methylenetetrahydrofolate reductase gene result in two enzyme isoforms. Mamm. Genome 2002, 13, 483–492.
- Frosst, P.; Blom, H.J.; Milos, R.; Goyette, P.; Sheppard, C.A.; Matthews, R.G.; Boers, G.J.; den Heijer, M.; Kluijtmans, L.A.; van den Heuvel, L.P.; et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995, 10, 111–113.
- Swanson, D.A.; Liu, M.L.; Baker, P.J.; Garrett, L.; Stitzel, M.; Wu, J.; Harris, M.; Banerjee, R.; Shane, B.; Brody, L.C. Targeted disruption of the methionine synthase gene in mice. Mol. Cell. Biol. 2001, 21, 1058–1065.
- Mentch, S.J.; Locasale, J.W. One-carbon metabolism and epigenetics: Understanding the specificity. Ann. N. Y. Acad. Sci. 2016, 1363, 91–98.
- Chiang, P.K.; Gordon, R.K.; Tal, J.; Zeng, G.C.; Doctor, B.P.; Pardhasaradhi, K.; McCann, P.P. S-Adenosylmethionine and methylation. FASEB J. 1996, 10, 471–480.
- Selhub, J. Homocysteine metabolism. Annu. Rev. Nutr. 1999, 19, 217–246.
- Mudd, S.H.; Uhlendorf, B.W.; Freeman, J.M.; Finkelstein, J.D.; Shih, V.E. Homocystinuria associated with decreased methylenetetrahydrofolate reductase activity. Biochem. Biophys. Res. Commun. 1972, 46, 905–912.
- Kang, S.S.; Zhou, J.; Wong, P.W.; Kowalisyn, J.; Strokosch, G. Intermediate homocysteinemia: A thermolabile variant of methylenetetrahydrofolate reductase. Am. J. Hum. Genet. 1988, 43, 414–421.
- Kang, S.S.; Wong, P.W.; Bock, H.G.; Horwitz, A.; Grix, A. Intermediate hyperhomocysteinemia resulting from compound heterozygosity of methylenetetrahydrofolate reductase mutations. Am. J. Hum. Genet. 1991, 48, 546–551.
- Raza, S.T.; Abbas, S.; Ahmed, F.; Fatima, J.; Zaidi, Z.H.; Mahdi, F. Association of MTHFR and PPARγ2 gene polymorphisms in relation to type 2 diabetes mellitus cases among north Indian population. Gene 2012, 511, 375–379.
- Nefic, H.; Mackic-Djurovic, M.; Eminovic, I. The Frequency of the 677C>T and 1298A>C Polymorphisms in the Methylenetetrahydrofolate Reductase (MTHFR) Gene in the Population. Med. Arch. 2018, 72, 164–169.
- Yadav, U.; Kumar, P.; Rai, V. Global prevalence of MTHFR C677T gene polymorphism: A meta-analysis of population based studies. Indian J. Clin. Biochem. 2014, 29, 123.
- van der Put, N.M.; Gabreëls, F.; Stevens, E.M.; Smeitink, J.A.; Trijbels, F.J.; Eskes, T.K.; van den Heuvel, L.P.; Blom, H.J. A second common mutation in the methylenetetrahydrofolate reductase gene: An additional risk factor for neural-tube defects? Am. J. Hum. Genet. 1998, 62, 1044–1051.
- Matthews, R.G.; Daubner, S.C. Modulation of methylenetetrahydrofolate reductase activity by S-adenosylmethionine and by dihydrofolate and its polyglutamate analogues. Adv. Enzym. Regul. 1982, 20, 123–131.
- Sumner, J.; Jencks, D.A.; Khani, S.; Matthews, R.G. Photoaffinity labeling of methylenetetrahydrofolate reductase with 8-azido-S-adenosylmethionine. J. Biol. Chem. 1986, 261, 7697–7700.
- Yamada, K.; Chen, Z.; Rozen, R.; Matthews, R.G. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc. Natl. Acad. Sci. USA 2001, 98, 14853–14858.
- Froese, D.S.; Kopec, J.; Rembeza, E.; Bezerra, G.A.; Oberholzer, A.E.; Suormala, T.; Lutz, S.; Chalk, R.; Borkowska, O.; Baumgartner, M.R.; et al. Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition. Nat. Commun. 2018, 9, 2261.
- Yamada, K.; Strahler, J.R.; Andrews, P.C.; Matthews, R.G. Regulation of human methylenetetrahydrofolate reductase by phosphorylation. Proc. Natl. Acad. Sci. USA 2005, 102, 10454–10459.
- Zheng, Y.; Ramsamooj, S.; Li, Q.; Johnson, J.L.; Yaron, T.M.; Sharra, K.; Cantley, L.C. Regulation of folate and methionine metabolism by multisite phosphorylation of human methylenetetrahydrofolate reductase. Sci. Rep. 2019, 9, 4190.
More
Information
Subjects:
Medicine, Research & Experimental
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
482
Revisions:
2 times
(View History)
Update Date:
09 Jan 2024
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




