Infectious diseases limit productivity and result in significant economic losses in each sector. Transboundary animal diseases (TADs) are economically important, have global reach, and require management. TADs can have significant implications for food security. Food-borne pathogens comprise microorganisms such as bacteria, viruses, and fungi, as well as parasites that cause food spoilage and infection. Food-borne pathogens are a major threat to food safety, as they can cause human diseases if animal products infected with toxins are consumed. The emergence of diseases stems from intricate interactions between microbes and humans, often influenced by a variety of complex factors. Key contributors to disease emergence include microbial adaptation and change, ecological shifts, human demographics and behavior, advancements in technology and healthcare, travel, trade, and industrial activities, breakdowns in public health measures, and varying levels of susceptibility to infection.
1. Zoonotic Viral Pathogens
Exposure to meat from an infected animal can lead to zoonotic food-borne infections. Although this type of transmission is a valid concern, it is the least common method of virus transmission
[1]. Most zoonotic infections are primarily acquired through direct contact with an infected animal, which can result in cutaneous lesions at the point of contact. However, there is also evidence to suggest that certain cases have occurred through mucosal pathways or after contact with infected surfaces (fomites). Animals, particularly wild animals, are thought to be the source of more than 70% of all new illnesses in humans
[2]. In recent decades, the chikungunya virus, human immunodeficiency virus type 1, Ebola virus, hantavirus pulmonary syndrome virus, Hendra virus, Nipah virus, severe acute respiratory syndrome (SARS), and coronavirus (COVID-19) are examples of viruses that have caused emergent diseases in humans
[3][4][5]. The transportation of companion animals that are afflicted could cause the poxvirus to be released into a new habitat, which is a probable scenario for a future outbreak. The current understanding of the epidemiology of poxvirus points to the need for more effective detection and management of these infections. At least three genera of poxviruses, including orthopoxvirus and parapoxvirus, contain zoonotic poxviruses. Food-related incidents have been linked to the transmission of (SARS), monkeypox, norovirus and the Ebola virus
[6]. Foodborne viruses enter the host organism through the gastrointestinal tract and reproduce in the intestinal tract before spreading throughout the body via the lymph nodes. Thus, the pathogenicity of the entering virus is influenced by its survival in the harsh, acidic environment of the stomach and proteolytic enzymes in the intestinal system.
The ‘gold standard’ for the detection of the majority of viruses is the polymerase chain reaction (PCR) method, which is rapid (a few hours to provide results) and highly specific. Real-time reverse transcriptase-PCR (RT-PCR) and qRT-PCR, which enable viral RNA detection, are of great interest because of their effectiveness needed for the efficient prevention of infection spreading. Both methods are equally effective in the detection of bacterial pathogens
[7].
Hepatitis E virus (HEV) is prevalent on domestic swine farms around the world, and it can infect pigs of all ages. As a result, the majority of foodborne HEV outbreaks have been linked to pork liver and pork liver-containing products
[8][9]. HEV outbreaks have also been linked to other foods, including cow milk and the meat of wild animals. HEV typically manifests as delimiting acute hepatitis in high-risk populations. Moreover, persistent hepatitis and extrahepatic symptoms may occur
[8]. The diagnosis of HEV infection can be performed through direct detection of the viral biomarker or indirect detection based on the host’s immune response to HEV. The direct tests detect parts of the viral particles, such as HEV ribonucleic acid (RNA) or viral capsid antigens. They usually have high specificity and low sensitivity
[10]. In contrast, the indirect tests targeting anti-HEV antibodies in blood have high sensitivity but low specificity. Consequently, diagnostic testing for suspected patients is usually performed by combining serology and nucleic acid amplification testing. The new generation of available HEV diagnostic tests is of advanced performance, but tests are still not standardized, and in many parts of the world, no diagnostic kit for commercial usage is available. This prevents the efficient control of the spread of the disease. Aquatic wild birds constitute the main reservoir for avian influenza virus (AIV). These viruses represent a global threat to animal health and the poultry industry and may cause a zoonotic infection that has effectively transcended the host organism threshold to infect humans
[11]. There is particularly high concern for pandemic emergences, which may have serious consequences on human health and cause enormous economic losses. AIVs are divided into low and highly pathogenic strains regarding their pathogenicity in chicken. The highly pathogenic AIV (HPAIV) causes systemic infections and results in a high mortality rate. The infection of poultry with low pathogenic avian viruses (LPAIV) generally leads to mild clinical signs, while in waterfowl species, often no clinical signs are invoked. Certain subtypes can change from low to high pathogenicity
[12], as in the case for LPAIV strains of the H5 and H7 subtypes, which acquired a highly pathogenic phenotype during infections in avian species. HPAIVs have killed both domestic and wild birds and have led to the destruction of hundreds of millions of domestic birds (e.g., around 30 million chickens were killed in The Netherlands, while 0.8 million were killed in France during the AIV outbreaks in 2003 and 2017, respectively)
[13]. South Korea faced an H5N6/H5N8 outbreak, which paralyzed its poultry industry; 20 million birds have been killed in January 2017. Such pandemic HPAIVs are difficult to control because no tool for in-field diagnostics is available. Samples collected by veterinarians at farms are first transported to authorized laboratories where the diagnosis of an HPAIV strain takes at least a whole day. To contain and eradicate zoonotic influenza viruses, researchers must not only conduct strategic virus surveillance in both animal and human populations but also gain a better understanding of the obstacles that a virus must overcome to cross the species barrier and infect humans. The influenza pandemic of 1918-1919, also known as the Spanish flu, caused widespread sickness and resulted in an estimated 40 million deaths worldwide. Existing studies have revealed that the pandemic virus contained genes that were derived from avian-like influenza virus strains. This virus is the common ancestor of both human and classical swine H1N1 influenza viruses. Pigs are believed to have played a role in this process, as they can be infected with both avian and human virus strains, which has resulted in various reassortants being isolated from them
[14].
Standard diagnostic methods based on virus propagation and isolation from embryonated chicken eggs are effective and sensitive, but they are time consuming and require complex procedures for sample collection and handling. Molecular methods based on RT-PCR need extracted genetic material. Portable later-flow devices proposed for some viral disease diagnostics are not multiplex and lack sensitivity
[15]. There is strong interest in developing new point-of-care biosensing systems for the early detection of viral diseases with high sensitivity and specificity. Moreover, a new version of the RT-PCR assay has been developed in accordance with the One Health program to meet the criteria of multi-species origin IAV detection. The matrix protein area is thought to be the most important aspect for detecting all AIV subtypes; however, given the amount of genetic drift that occurs over time, changes to this matrix protein region have added novelty to this assay
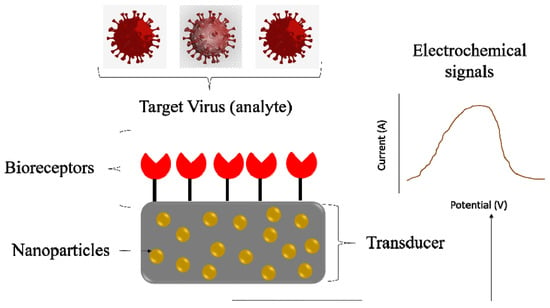
[16]. Rapid and robust virus detection methods could significantly help in future pandemics. Biosensors for virus detection based on electrochemical, plasmonic, and optical signals make them ideal platforms for virus detection (
Figure 1). Combining such portable devices with suitable nanomaterials can enhance the portfolio of available diagnostic kits. Biosensors’ sensitivity and selectivity are usually connected with the recognition element (antibody, DNA probe, aptamer) changing electronic or optical properties in the presence of the targeted virus. Biosensor strategies can be readily adopted for the detection of new emerging zoonotic viruses.
Figure 1. Biosensors based electrochemical signal for virus detection.
2. Antimicrobial Resistant Pathogens
The rise of antimicrobial resistance (AMR) is due to the misuse and overuse of antibiotics in both humans and veterinary and agricultural practices
[17]. The World Organisation for Animal Health (OIE), along with the Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO), consider antibiotic resistance to be a major priority. Managing this issue requires coordinated and concerted efforts across multiple sectors, including animal and agricultural production, food processing, human health, and the environment. The non-rational use of veterinary antibiotics may result in the pressure selection of resistant pathogens. While increasing AMR awareness is critical, new antibiotics and therapy techniques must also be developed. The One Health platform raises public awareness about AMR by implementing a dual AMR track. To gain a new perspective, an algorithm was developed as a tool to quickly assess the potential for a new or emerging livestock disease to harm humans through the consumption or handling of meat products, so that the risks and uncertainties can be understood, and appropriate precautions and policies can be enacted. The One Health systematic method of assessing AMR will aid in a better understanding of antibiotic resistance
[18]. The surveillance of AMR, legislative reforms, new economic models, diagnostics and detections, and alternative techniques to combat resistant infections should all be considered for a rapid response to this issue. Antibiotic stewardship refers to efforts to enhance the use of various antibiotics to prevent needless antibiotic use
[18]. There is a variety of options for the use of antibiotics, including the use of bacteriophages and immune modulators
[19]. The sequence of events from the onset of the disease in cattle to the discharge of the causative agent from an infected animal, the contamination of fresh meat, and possible harmful consequences in humans following contact with meat was developed using an algorithm. The concept of One Health has long been a component of human civilization. The One Health approach can grasp the interconnectedness and inherent complexities of human and animal health and the environment by addressing their relationship. This extraordinary method may inspire scientists to develop new fields of research to generate innovative ideas. Science has demonstrated its ability to successfully integrate all sectors to identify a path for detecting emerging diseases and pathogens, as well as developing novel therapeutic procedures
[20][21].
The prevalence of organisms exhibiting AMR, especially resistance to multiple antibiotics, shows large variations in the percentages of AMR depending on the microorganism, antimicrobial agent, and geographical region
[22]. Most of the ESKAPE pathogens (Enterococcus faecium, S. aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa, and Enterobacter species) are multidrug resistant isolates, and one of the greatest challenges in clinical practice. Initially, the ESKAPE bacteria surveillance was focused on healthcare-associated infections. Today, the increasing AMR awareness of ESKAPE strains has led to extensive investigations in various ecosystems. The presence of multi-resistant ESKAPE strains carrying mobile genetic elements and gene cassettes encoding resistance to antibiotics or biocides in the water cycle is not demonstrated. Both freshwater and marine systems act as a sink for ESKAPE bacteria that enter aquatic systems through treated and untreated sewage, hospital waste, and agricultural run-off
[23]. Water contamination with AMR bacteria, especially water co-contaminated with antibiotic residues, may induce resistance in autochthonous bacteria through horizontal gene transfer as well as through spontaneous de novo point mutations, which can change the cellular targets of antibiotics or the expression of resistant genes, leading to increased antibiotic resistant of the bacteria
[24]. In horizontal gene transfer, genetic material can transfer between related or unrelated species via mobile elements. Given the potential for transmission and environmental dissemination, ESKAPE infections, which are well known for their antibiotic resistance in human healthcare, indirectly connect with the One Health paradigm. Antibiotic-resistant bacteria can be identified in animals and the environment, while being largely linked with humans, demonstrating how interrelated humans, animals, and environmental health are
[25][26][27].