
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Pramod Chaudhary | -- | 2563 | 2023-12-22 09:40:41 |
Video Upload Options
Skin cancer, a malignant neoplasm originating from skin cell types including keratinocytes, melanocytes, and sweat glands, comprises three primary forms: basal cell carcinoma (BCC), squamous cell carcinoma (SCC), and malignant melanoma (MM). BCC and SCC, while constituting the most prevalent categories of skin cancer, are generally considered less aggressive compared to MM. Notably, MM possesses a greater capacity for invasiveness, enabling infiltration into adjacent tissues and dissemination via both the circulatory and lymphatic systems. Risk factors associated with skin cancer encompass ultraviolet (UV) radiation exposure, fair skin complexion, a history of sunburn incidents, genetic predisposition, immunosuppressive conditions, and exposure to environmental carcinogens. Early detection of skin cancer is of paramount importance to optimize treatment outcomes and preclude the progression of disease, either locally or to distant sites. In pursuit of this objective, numerous computer-aided diagnosis (CAD) systems have been developed. Hyperspectral imaging (HSI), distinguished by its capacity to capture information spanning the electromagnetic spectrum, surpasses conventional RGB imaging, which relies solely on three color channels.
1. Introduction
2. Methods of Skin Cancer Detection
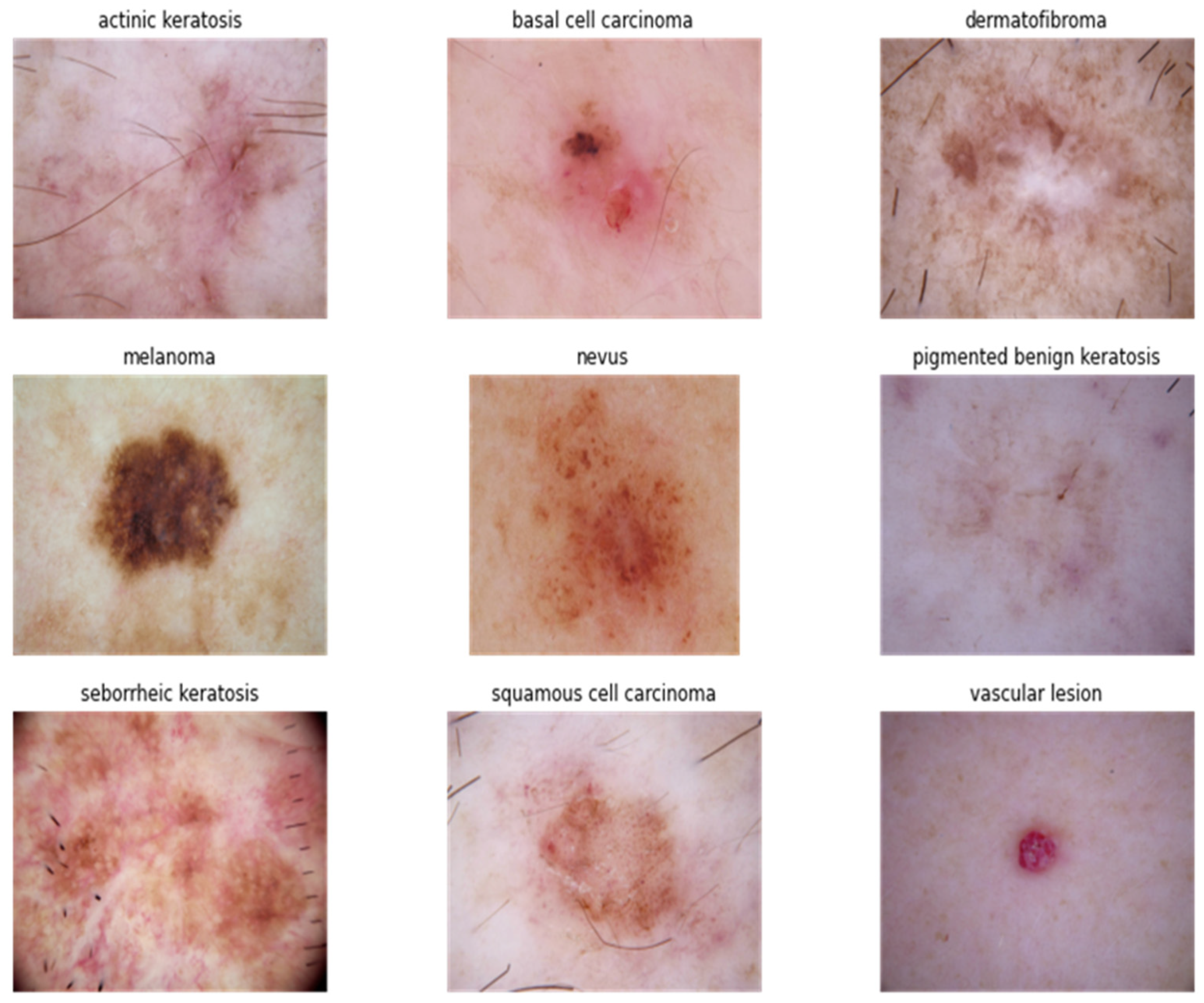
3. Computer-Aided Detection Methods Using Hyperspectral Imaging Engineering to Detect Skin Cancer
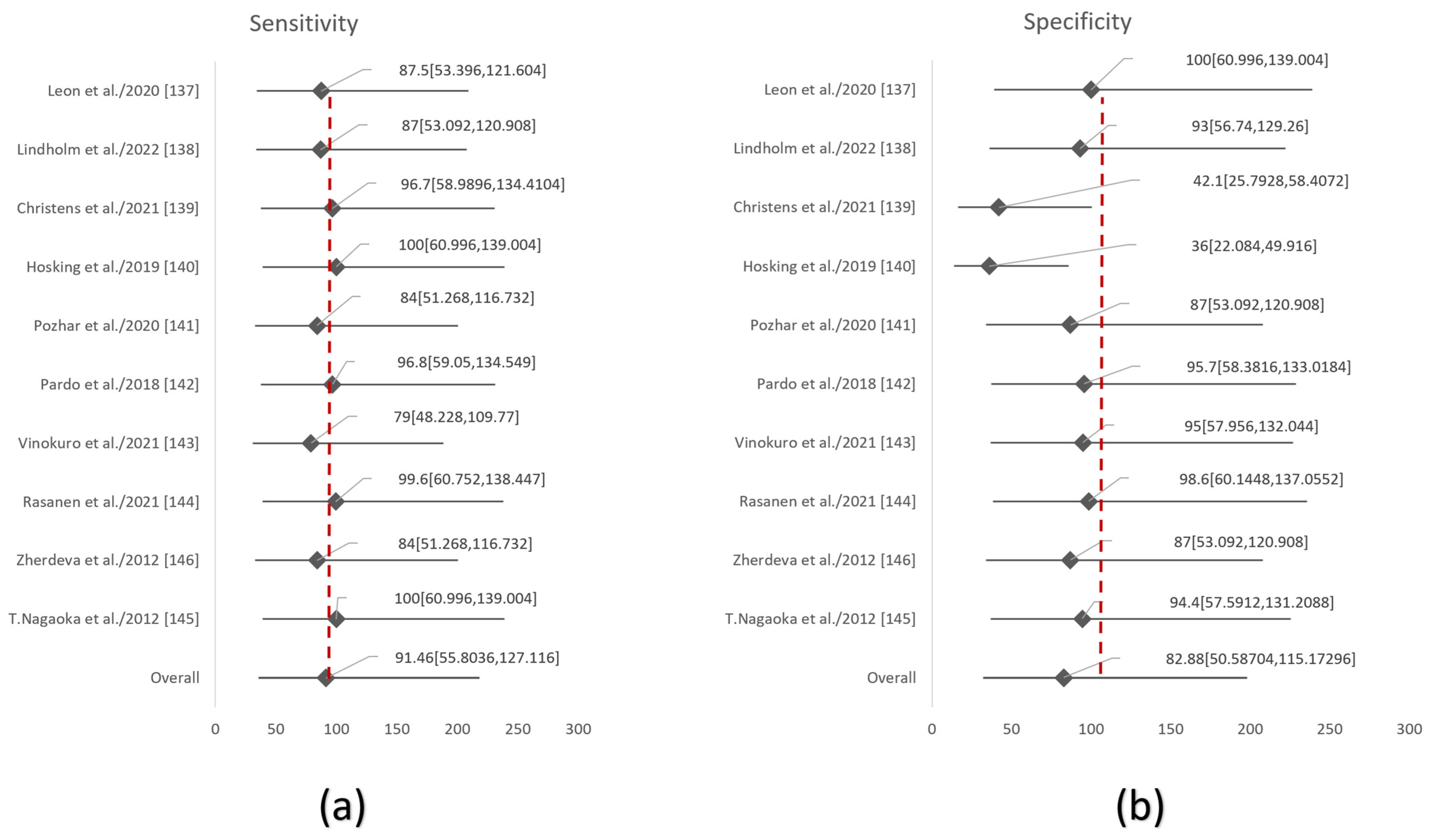
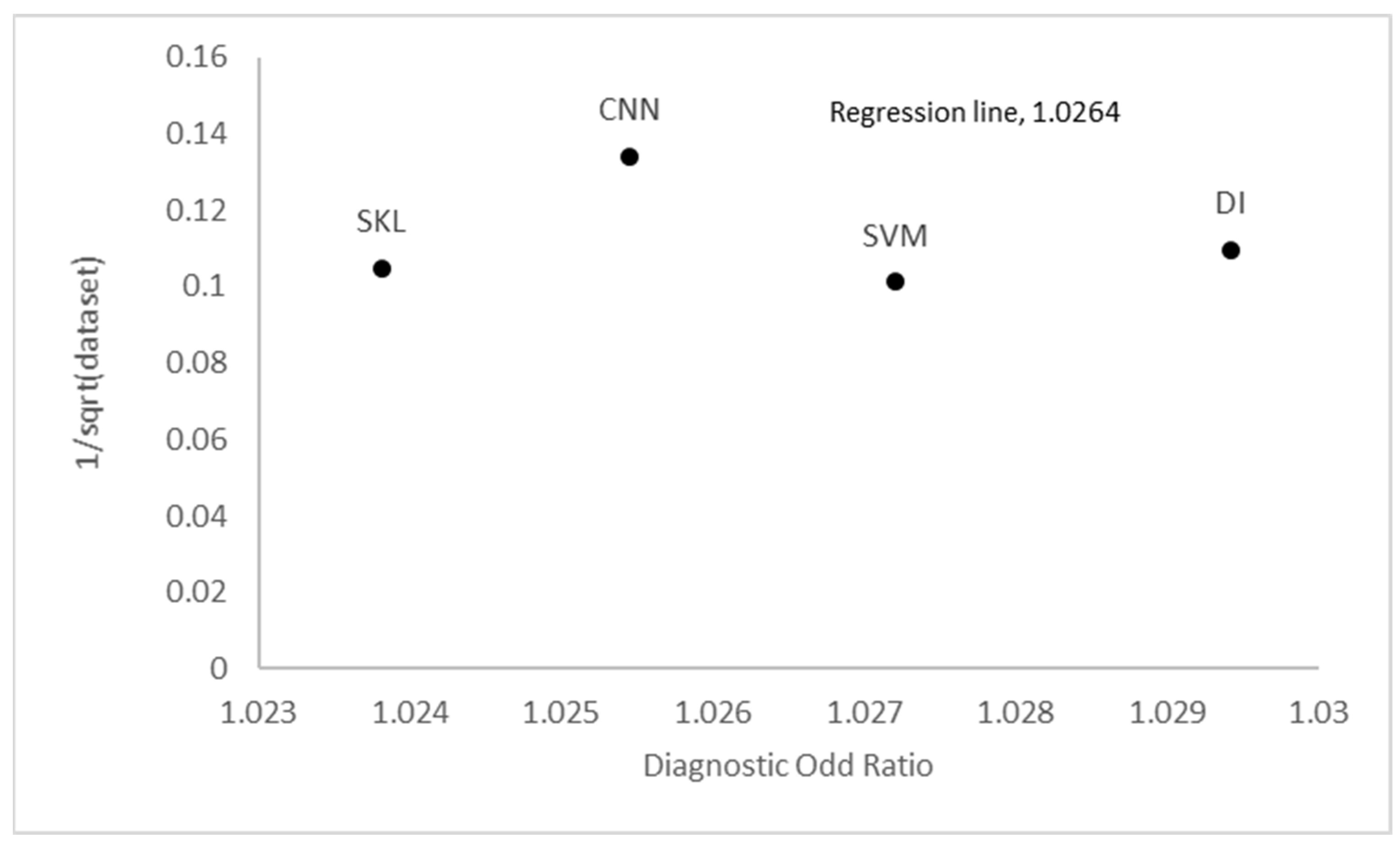
References
- Gupta, A.K.; Bharadwaj, M.; Mehrotra, R. Skin cancer concerns in people of color: Risk factors and prevention. Asian Pac. J. Cancer Prev. APJCP 2016, 17, 5257.
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2021, 71, 209–249.
- Leiter, U.; Keim, U.; Garbe, C. Epidemiology of skin cancer: Update 2019. In Sunlight, Vitamin D and Skin Cancer; Springer: Cham, Switzerland, 2020; pp. 123–139.
- Fahradyan, A.; Howell, A.C.; Wolfswinkel, E.M.; Tsuha, M.; Sheth, P.; Wong, A.K. Updates on the management of non-melanoma skin cancer (NMSC). Healthcare 2017, 5, 82.
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Mathers, C.; Parkin, D.M.; Piñeros, M.; Znaor, A.; Bray, F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int. J. Cancer 2019, 144, 1941–1953.
- Khan, M.N.; Wang, Q.; Idrees, B.S.; Xiangli, W.; Teng, G.; Cui, X.; Zhao, Z.; Wei, K.; Abrar, M. A review on laser-induced breakdown spectroscopy in different cancers diagnosis and classification. Front. Phys. 2022, 10, 10.
- Stevens, V.W.; Stenehjem, D.D.; Patterson, O.V.; Kamauu, A.W.; Yim, Y.M.; Morlock, R.J.; DuVall, S.L. Characterization and survival of patients with metastatic basal cell carcinoma in the Department of Veterans Affairs: A retrospective electronic health record review. Arch. Dermatol. Res. 2018, 310, 505–513.
- Stiegel, E.; Lam, C.; Schowalter, M.; Somani, A.-K.; Lucas, J.; Poblete-Lopez, C. Correlation between original biopsy pathology and Mohs intraoperative pathology. Dermatol. Surg. 2018, 44, 193–197.
- Khazaei, Z.; Ghorat, F.; Jarrahi, A.; Adineh, H.; Sohrabivafa, M.; Goodarzi, E. Global incidence and mortality of skin cancer by histological subtype and its relationship with the human development index (HDI)—An ecology study in 2018. World Cancer Res. J. 2019, 6, e13.
- Ciążyńska, M.; Kamińska-Winciorek, G.; Lange, D.; Lewandowski, B.; Reich, A.; Sławińska, M.; Pabianek, M.; Szczepaniak, K.; Hankiewicz, A.; Ułańska, M. The incidence and clinical analysis of non-melanoma skin cancer. Sci. Rep. 2021, 11, 4337.
- Popescu, D.; El-Khatib, M.; El-Khatib, H.; Ichim, L. New Trends in Melanoma Detection Using Neural Networks: A Systematic Review. Sensors 2022, 22, 496.
- Ahlgrimm-Siess, V.; Laimer, M.; Arzberger, E.; Hofmann-Wellenhof, R. New diagnostics for melanoma detection: From artificial intelligence to RNA microarrays. Future Oncol. 2012, 8, 819–827.
- Narayanan, D.L.; Saladi, R.N.; Fox, J.L. Ultraviolet radiation and skin cancer. Int. J. Dermatol. 2010, 49, 978–986.
- Federico, M.B. SDG 3 Good Health and Well-Being. In Actioning the Global Goals for Local Impact; Springer: Berlin/Heidelberg, Germany, 2020; pp. 39–55.
- Umar, S.A.; Tasduq, S.A. Ozone Layer Depletion and Emerging Public Health Concerns-An Update on Epidemiological Perspective of the Ambivalent Effects of Ultraviolet Radiation Exposure. Front. Oncol. 2022, 12, 866733.
- Lin, T.-C.; Lee, H.-C. Skin cancer dermoscopy images classification with meta data via deep learning ensemble. In Proceedings of the 2020 International Computer Symposium (ICS), Tainan, Taiwan, 17–19 December 2020; pp. 237–241.
- Kim, H.S.; Kim, H.J.; Hong, E.S.; Kim, K.B.; Lee, J.D.; Kang, T.U.; Ahn, H.S. The incidence and survival of melanoma and nonmelanoma skin cancer in patients with vitiligo: A nationwide population-based matched cohort study in Korea. Br. J. Dermatol. 2020, 182, 907–915.
- Giaquinto, A.N.; Miller, K.D.; Tossas, K.Y.; Winn, R.A.; Jemal, A.; Siegel, R.L. Cancer statistics for African American/Black People 2022. CA A Cancer J. Clin. 2022, 72, 202–229.
- Islami, F.; Guerra, C.E.; Minihan, A.; Yabroff, K.R.; Fedewa, S.A.; Sloan, K.; Wiedt, T.L.; Thomson, B.; Siegel, R.L.; Nargis, N. American Cancer Society’s report on the status of cancer disparities in the United States, 2021. CA A Cancer J. Clin. 2022, 72, 112–143.
- Iwagami, M.; Caplin, B.; Smeeth, L.; Tomlinson, L.A.; Nitsch, D. Clinical Codelist—Read Codes for Hypothyroidism; Data Collection; London School of Hygiene & Tropical Medicine: London, UK, 2018.
- Arnold, M.; Singh, D.; Laversanne, M.; Vignat, J.; Vaccarella, S.; Meheus, F.; Cust, A.E.; de Vries, E.; Whiteman, D.C.; Bray, F. Global burden of cutaneous melanoma in 2020 and projections to 2040. JAMA Dermatol. 2022, 158, 495–503.
- Di Carlo, V.; Stiller, C.A.; Eisemann, N.; Bordoni, A.; Matz, M.; Curado, M.P.; Daubisse-Marliac, L.; Valkov, M.; Bulliard, J.L.; Morrison, D. Does the morphology of cutaneous melanoma help explain the international differences in survival? Results from 1,578,482 adults diagnosed during 2000–2014 in 59 countries (CONCORD-3). Br. J. Dermatol. 2022, 187, 364–380.
- Perez, E.; Ventura, S. Multi-view Deep Neural Networks for multiclass skin lesion diagnosis. In Proceedings of the 2022 IEEE International Conference on Omni-layer Intelligent Systems (COINS), Barcelona, Spain, 1–3 August 2022; pp. 1–6.
- Siegel, R.; Ward, E.; Brawley, O.; Jemal, A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA A Cancer J. Clin. 2011, 61, 212–236.
- Gochi, A.; Orita, K.; Fuchimoto, S.; Tanaka, N.; Ogawa, N. The prognostic advantage of preoperative intratumoral injection of OK-432 for gastric cancer patients. Br. J. Cancer 2001, 84, 443–451.
- Ishihara, K.; Saida, T.; Otsuka, F.; Yamazaki, N. Statistical profiles of malignant melanoma and other skin cancers in Japan: 2007 update. Int. J. Clin. Oncol. 2008, 13, 33–41.
- Gloster, H.M., Jr.; Brodland, D.G. The epidemiology of skin cancer. Dermatol. Surg. 1996, 22, 217–226.
- Reilly, P.; Cosh, J.; Maddison, P.; Rasker, J.; Silman, A. Mortality and survival in rheumatoid arthritis: A 25 year prospective study of 100 patients. Ann. Rheum. Dis. 1990, 49, 363–369.
- Tseng, W.-P. Effects and dose-response relationships of skin cancer and blackfoot disease with arsenic. Environ. Health Perspect. 1977, 19, 109–119.
- Phadke, J.G. Survival pattern and cause of death in patients with multiple sclerosis: Results from an epidemiological survey in north east Scotland. J. Neurol. Neurosurg. Psychiatry 1987, 50, 523–531.
- Tsuchiya, K.; Aoyama, T.; Ju, M.; Atsumi, Y.; Kazama, K.; Numata, M.; Tamagawa, H.; Yukawa, N.; Rino, Y. A Case of Rectal Cancer with Brain and Skin Metastasis with Long-Term Survival Managed by Multidisciplinary Therapy. Gan Kagaku Ryoho 2022, 49, 1148–1150.
- Celebi, M.E.; Iyatomi, H.; Schaefer, G.; Stoecker, W.V. Lesion border detection in dermoscopy images. Comput. Med. Imaging Graph. 2009, 33, 148–153.
- Fernandes, S.L.; Chakraborty, B.; Gurupur, V.P.; Prabhu, G.A. Early skin cancer detection using computer aided diagnosis techniques. J. Integr. Des. Process. Sci. 2016, 20, 33–43.
- Adla, D.; Reddy, G.; Nayak, P.; Karuna, G. Deep learning-based computer aided diagnosis model for skin cancer detection and classification. Distrib. Parallel Databases 2022, 40, 717–736.
- Tsai, C.L.; Mukundan, A.; Chung, C.S.; Chen, Y.H.; Wang, Y.K.; Chen, T.H.; Tseng, Y.S.; Huang, C.W.; Wu, I.C.; Wang, H.C. Hyperspectral Imaging Combined with Artificial Intelligence in the Early Detection of Esophageal Cancer. Cancers 2021, 13, 4593.
- Xu, Z.; Sheykhahmad, F.R.; Ghadimi, N.; Razmjooy, N. Computer-aided diagnosis of skin cancer based on soft computing techniques. Open Med. 2020, 15, 860–871.
- Jaleel, J.A.; Salim, S.; Aswin, R. Computer aided detection of skin cancer. In Proceedings of the 2013 International Conference on Circuits, Power and Computing Technologies (ICCPCT), Nagercoil, India, 20–21 March 2013; pp. 1137–1142.
- Kumar, K.A.; Vanmathi, C. Optimization driven model and segmentation network for skin cancer detection. Comput. Electr. Eng. 2022, 103, 108359.
- Filali, Y.; EL Khoukhi, H.; Sabri, M.A.; Aarab, A. Efficient fusion of handcrafted and pre-trained CNNs features to classify melanoma skin cancer. Multimed. Tools Appl. 2020, 79, 31219–31238.
- Mohanty, S.P.; Kougianos, E. Biosensors: A tutorial review. Ieee Potentials 2006, 25, 35–40.
- Malibari, A.A.; Alzahrani, J.S.; Eltahir, M.M.; Malik, V.; Obayya, M.; Al Duhayyim, M.; Neto, A.V.L.; de Albuquerque, V.H.C. Optimal deep neural network-driven computer aided diagnosis model for skin cancer. Comput. Electr. Eng. 2022, 103, 108318.
- Bratchenko, I.A.; Bratchenko, L.A.; Moryatov, A.A.; Khristoforova, Y.A.; Artemyev, D.N.; Myakinin, O.O.; Orlov, A.E.; Kozlov, S.V.; Zakharov, V.P. In vivo diagnosis of skin cancer with a portable Raman spectroscopic device. Exp. Dermatol. 2021, 30, 652–663.
- Bohunicky, B.; Mousa, S. Biosensors: The new wave in cancer diagnosis. Nanotechnol. Sci. Appl. 2010, 4, 1–10.
- Keshavarz, A.; Vafapour, Z. Water-based terahertz metamaterial for skin cancer detection application. IEEE Sens. J. 2018, 19, 1519–1524.
- Lalitha, K.; Lakshmi, K. An overview on biosensors. Int. J. Pharm. Chem. Biol. Sci. 2017, 7, 293–302.
- Ashraf, R.; Afzal, S.; Rehman, A.U.; Gul, S.; Baber, J.; Bakhtyar, M.; Mehmood, I.; Song, O.-Y.; Maqsood, M. Region-of-interest based transfer learning assisted framework for skin cancer detection. IEEE Access 2020, 8, 147858–147871.
- Alheejawi, S.; Xu, H.; Berendt, R.; Jha, N.; Mandal, M. Novel lymph node segmentation and proliferation index measurement for skin melanoma biopsy images. Comput. Med. Imaging Graph. 2019, 73, 19–29.
- Vocaturo, E.; Perna, D.; Zumpano, E. Machine learning techniques for automated melanoma detection. In Proceedings of the 2019 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), San Diego, CA, USA, 18–21 November 2019; pp. 2310–2317.
- Rey-Barroso, L.; Peña-Gutiérrez, S.; Yáñez, C.; Burgos-Fernández, F.J.; Vilaseca, M.; Royo, S. Optical technologies for the improvement of skin cancer diagnosis: A review. Sensors 2021, 21, 252.
- Jiang, S.; Li, H.; Jin, Z. A visually interpretable deep learning framework for histopathological image-based skin cancer diagnosis. IEEE J. Biomed. Health Inform. 2021, 25, 1483–1494.
- Saba, T. Computer vision for microscopic skin cancer diagnosis using handcrafted and non-handcrafted features. Microsc. Res. Tech. 2021, 84, 1272–1283.
- Kumar, M.; Alshehri, M.; AlGhamdi, R.; Sharma, P.; Deep, V. A de-ann inspired skin cancer detection approach using fuzzy c-means clustering. Mob. Netw. Appl. 2020, 25, 1319–1329.
- Premaladha, J.; Ravichandran, K. Novel approaches for diagnosing melanoma skin lesions through supervised and deep learning algorithms. J. Med. Syst. 2016, 40, 96.
- Afifi, S.; GholamHosseini, H.; Sinha, R. SVM classifier on chip for melanoma detection. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. 2017, 2017, 270–274.
- Thiem, D.G.; Römer, P.; Blatt, S.; Al-Nawas, B.; Kämmerer, P.W. New Approach to the Old Challenge of Free Flap Monitoring—Hyperspectral Imaging Outperforms Clinical Assessment by Earlier Detection of Perfusion Failure. J. Pers. Med. 2021, 11, 1101.
- Ghamisi, P.; Rasti, B.; Yokoya, N.; Wang, Q.; Hofle, B.; Bruzzone, L.; Bovolo, F.; Chi, M.; Anders, K.; Gloaguen, R. Multisource and multitemporal data fusion in remote sensing: A comprehensive review of the state of the art. IEEE Geosci. Remote Sens. Mag. 2019, 7, 6–39.
- Li, Q.; He, X.; Wang, Y.; Liu, H.; Xu, D.; Guo, F. Review of spectral imaging technology in biomedical engineering: Achievements and challenges. J. Biomed. Opt. 2013, 18, 100901.
- Lu, G.; Fei, B. Medical hyperspectral imaging: A review. J. Biomed. Opt. 2014, 19, 010901.
- Zakian, C.M.; Pretty, I.A.; Ellwood, R. Near-infared hyperspectral imaging of teeth for dental caries detection. J. Biomed. Opt. 2009, 14, 064047.
- Tromberg, B.J.; Shah, N.; Lanning, R.; Cerussi, A.; Espinoza, J.; Pham, T.; Svaasand, L.; Butler, J. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia 2000, 2, 26–40.
- Cerussi, A.E.; Berger, A.J.; Bevilacqua, F.; Shah, N.; Jakubowski, D.; Butler, J.; Holcombe, R.F.; Tromberg, B.J. Sources of absorption and scattering contrast for near-infrared optical mammography. Acad. Radiol. 2001, 8, 211–218.
- Bi, D.; Zhu, D.; Sheykhahmad, F.R.; Qiao, M. Computer-aided skin cancer diagnosis based on a New meta-heuristic algorithm combined with support vector method. Biomed. Signal Process. Control 2021, 68, 102631.
- Barducci, A.; Guzzi, D.; Marcoionni, P.; Pippi, I. Aerospace wetland monitoring by hyperspectral imaging sensors: A case study in the coastal zone of San Rossore Natural Park. J. Environ. Manag. 2009, 90, 2278–2286.
- Sun, D.-W. Hyperspectral Imaging for Food Quality Analysis and Control; Elsevier: Amsterdam, The Netherlands, 2010.
- Lu, B.; Dao, P.D.; Liu, J.; He, Y.; Shang, J. Recent advances of hyperspectral imaging technology and applications in agriculture. Remote Sens. 2020, 12, 2659.
- Fei, B. Hyperspectral imaging in medical applications. In Data Handling in Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; Volume 32, pp. 523–565.
- Lee, C.-H.; Mukundan, A.; Chang, S.-C.; Wang, Y.-L.; Lu, S.-H.; Huang, Y.-C.; Wang, H.-C. Comparative Analysis of Stress and Deformation between One-Fenced and Three-Fenced Dental Implants Using Finite Element Analysis. J. Clin. Med. 2021, 10, 3986.
- Hege, E.K.; O’Connell, D.; Johnson, W.; Basty, S.; Dereniak, E.L. Hyperspectral imaging for astronomy and space surveillance. Imaging Spectrom. IX 2004, 5159, 380–391.
- Courtenay, L.A.; González-Aguilera, D.; Lagüela, S.; Del Pozo, S.; Ruiz-Mendez, C.; Barbero-García, I.; Román-Curto, C.; Cañueto, J.; Santos-Durán, C.; Cardeñoso-Álvarez, M.E. Hyperspectral imaging and robust statistics in non-melanoma skin cancer analysis. Biomed. Opt. Express 2021, 12, 5107–5127.
- Tsai, T.-J.; Mukundan, A.; Chi, Y.-S.; Tsao, Y.-M.; Wang, Y.-K.; Chen, T.-H.; Wu, I.-C.; Huang, C.-W.; Wang, H.-C. Intelligent Identification of Early Esophageal Cancer by Band-Selective Hyperspectral Imaging. Cancers 2022, 14, 4292.
- Fang, Y.-J.; Mukundan, A.; Tsao, Y.-M.; Huang, C.-W.; Wang, H.-C. Identification of Early Esophageal Cancer by Semantic Segmentation. J. Pers. Med. 2022, 12, 1204.
- Aboughaleb, I.H.; Aref, M.H.; El-Sharkawy, Y.H. Hyperspectral imaging for diagnosis and detection of ex-vivo breast cancer. Photodiagnosis Photodyn. Ther. 2020, 31, 101922.
- Liu, H.; Yu, T.; Hu, B.; Hou, X.; Zhang, Z.; Liu, X.; Liu, J.; Wang, X.; Zhong, J.; Tan, Z. Uav-borne hyperspectral imaging remote sensing system based on acousto-optic tunable filter for water quality monitoring. Remote Sens. 2021, 13, 4069.
- Liu, H.; Bruning, B.; Garnett, T.; Berger, B. Hyperspectral imaging and 3D technologies for plant phenotyping: From satellite to close-range sensing. Comput. Electron. Agric. 2020, 175, 105621.
- Shrestha, S.; Knapič, M.; Žibrat, U.; Deleuran, L.C.; Gislum, R. Single seed near-infrared hyperspectral imaging in determining tomato (Solanum lycopersicum L.) seed quality in association with multivariate data analysis. Sens. Actuators B Chem. 2016, 237, 1027–1034.
- Wu, N.; Liu, F.; Meng, F.; Li, M.; Zhang, C.; He, Y. Rapid and accurate varieties classification of different crop seeds under sample-limited condition based on hyperspectral imaging and deep transfer learning. Front. Bioeng. Biotechnol. 2021, 9, 696292.
- Hsiao, Y.-P.; Mukundan, A.; Chen, W.-C.; Wu, M.-T.; Hsieh, S.-C.; Wang, H.-C. Design of a Lab-On-Chip for Cancer Cell Detection through Impedance and Photoelectrochemical Response Analysis. Biosensors 2022, 12, 405.
- Stuart, M.B.; McGonigle, A.J.; Willmott, J.R. Hyperspectral imaging in environmental monitoring: A review of recent developments and technological advances in compact field deployable systems. Sensors 2019, 19, 3071.
- Chen, C.-W.; Tseng, Y.-S.; Mukundan, A.; Wang, H.-C. Air Pollution: Sensitive Detection of PM2.5 and PM10 Concentration Using Hyperspectral Imaging. Appl. Sci. 2021, 11, 4543.
- Huang, S.-Y.; Mukundan, A.; Tsao, Y.-M.; Kim, Y.; Lin, F.-C.; Wang, H.-C. Recent Advances in Counterfeit Art, Document, Photo, Hologram, and Currency Detection Using Hyperspectral Imaging. Sensors 2022, 22, 7308.
- Mukundan, A.; Tsao, Y.-M.; Lin, F.-C.; Wang, H.-C. Portable and low-cost hologram verification module using a snapshot-based hyperspectral imaging algorithm. Sci. Rep. 2022, 12, 18475.
- Mukundan, A.; Wang, H.-C.; Tsao, Y.-M. A Novel Multipurpose Snapshot Hyperspectral Imager used to Verify Security Hologram. In Proceedings of the 2022 International Conference on Engineering and Emerging Technologies (ICEET), Kuala Lumpur, Malaysia, 27–28 October 2022; pp. 1–3.
- Mukundan, A.; Tsao, Y.-M.; Cheng, W.-M.; Lin, F.-C.; Wang, H.-C. Automatic Counterfeit Currency Detection Using a Novel Snapshot Hyperspectral Imaging Algorithm. Sensors 2023, 23, 2026.
- Hamilton, S.J.; Lodder, R.A. Hyperspectral imaging technology for pharmaceutical analysis. Biomed. Nanotechnol. Archit. Appl. 2002, 4626, 136–147.
- Chang, L.; Zhang, M.; Li, W. A coarse-to-fine approach for medical hyperspectral image classification with sparse representation. AOPC 2017 Opt. Spectrosc. Imaging 2017, 10461, 136–144.
- Yang, K.-Y.; Fang, Y.-J.; Karmakar, R.; Mukundan, A.; Tsao, Y.-M.; Huang, C.-W.; Wang, H.-C. Assessment of Narrow Band Imaging Algorithm for Video Capsule Endoscopy Based on Decorrelated Color Space for Esophageal Cancer. Cancers 2023, 15, 4715.
- de la Ossa, M.Á.F.; Amigo, J.M.; García-Ruiz, C. Detection of residues from explosive manipulation by near infrared hyperspectral imaging: A promising forensic tool. Forensic Sci. Int. 2014, 242, 228–235.
- Favreau, P.F.; Hernandez, C.; Lindsey, A.S.; Alvarez, D.F.; Rich, T.C.; Prabhat, P.; Leavesley, S.J. Thin-film tunable filters for hyperspectral fluorescence microscopy. J. Biomed. Opt. 2013, 19, 011017.
- Xu, D.; Ni, G.; Jiang, T.; Jiang, L.; Chi, M. Integration of field work and hyperspectral data for oil and gas exploration. In Proceedings of the 2007 IEEE International Geoscience and Remote Sensing Symposium, Barcelona, Spain, 23–28 July 2007; pp. 3194–3197.
- Gowen, A.A.; Feng, Y.; Gaston, E.; Valdramidis, V. Recent applications of hyperspectral imaging in microbiology. Talanta 2015, 137, 43–54.
- Wang, Y.; Hu, X.; Hou, Z.; Ning, J.; Zhang, Z. Discrimination of nitrogen fertilizer levels of tea plant (Camellia sinensis) based on hyperspectral imaging. J. Sci. Food Agric. 2018, 98, 4659–4664.
- Huang, S.; Wang, L.; Chen, W.; Lin, D.; Huang, L.; Wu, S.; Feng, S.; Chen, R. Non-invasive optical detection of esophagus cancer based on urine surface-enhanced Raman spectroscopy. In Proceedings of the Twelfth International Conference on Photonics and Imaging in Biology and Medicine (PIBM 2014), Wuhan, China, 17 September 2014; pp. 537–542.
- Zabalza, J.; Ren, J.; Wang, Z.; Marshall, S.; Wang, J. Singular spectrum analysis for effective feature extraction in hyperspectral imaging. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1886–1890.
- Fabelo, H.; Ortega, S.; Lazcano, R.; Madroñal, D.; Callicó, G.M.; Juárez, E.; Salvador, R.; Bulters, D.; Bulstrode, H.; Szolna, A. An intraoperative visualization system using hyperspectral imaging to aid in brain tumor delineation. Sensors 2018, 18, 430.
- More, S.S.; Beach, J.M.; McClelland, C.; Mokhtarzadeh, A.; Vince, R. In vivo assessment of retinal biomarkers by hyperspectral imaging: Early detection of Alzheimer’s disease. ACS Chem. Neurosci. 2019, 10, 4492–4501.
- Chang, C.-I.; Wu, C.-C.; Liu, K.-H.; Chen, H.-M.; Chen, C.C.-C.; Wen, C.-H. Progressive band processing of linear spectral unmixing for hyperspectral imagery. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 8, 2583–2597.
- Akbari, H.; Halig, L.V.; Zhang, H.; Wang, D.; Chen, Z.G.; Fei, B. Detection of cancer metastasis using a novel macroscopic hyperspectral method. In Proceedings of the Medical Imaging 2012: Biomedical Applications in Molecular, Structural, and Functional Imaging, San Diego, CA, USA, 14 April 2012; pp. 299–305.
- Senan, E.M.; Jadhav, M.E. Classification of dermoscopy images for early detection of skin cancer—A review. Int. J. Comput. Appl. 2019, 975, 8887.
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-level classification of skin cancer with deep neural networks. Nature 2017, 542, 115–118.
- Ortega, S.; Fabelo, H.; Iakovidis, D.K.; Koulaouzidis, A.; Callico, G.M. Use of hyperspectral/multispectral imaging in gastroenterology. Shedding some–different–light into the dark. J. Clin. Med. 2019, 8, 36.
- Nachbar, F.; Stolz, W.; Merkle, T.; Cognetta, A.B.; Vogt, T.; Landthaler, M.; Bilek, P.; Braun-Falco, O.; Plewig, G. The ABCD rule of dermatoscopy: High prospective value in the diagnosis of doubtful melanocytic skin lesions. J. Am. Acad. Dermatol. 1994, 30, 551–559.
- Tsao, H.; Olazagasti, J.M.; Cordoro, K.M.; Brewer, J.D.; Taylor, S.C.; Bordeaux, J.S.; Chren, M.-M.; Sober, A.J.; Tegeler, C.; Bhushan, R. Early detection of melanoma: Reviewing the ABCDEs. J. Am. Acad. Dermatol. 2015, 72, 717–723.
- Peters, M.S.; Winkelmann, R.K. The biopsy. Dermatol. Clin. 1984, 2, 209–217.
- Wollina, U.; Burroni, M.; Torricelli, R.; Gilardi, S.; Dell’Eva, G.; Helm, C.; Bardey, W. Digital dermoscopy in clinical practise: A three-centre analysis. Ski. Res. Technol. 2007, 13, 133–142.
- Tudor, A.; Feldman, J.; Diamandis, C. Why the ABCDE Rule Is Not Helpful but Dangerous in Skin Cancer Prevention. Zenodo, 27 November 2021. Available online: https://www.scanoma.com/blog/why-the-abcde-rule-is-not-helpful-but-dangerous-in-skin-cancer-prevention (accessed on 26 November 2023).
- Fabelo, H.; Melián, V.; Martínez, B.; Beltrán, P.; Ortega, S.; Marrero, M.; Callicó, G.M.; Sarmiento, R.; Castaño, I.; Carretero, G. Dermatologic hyperspectral imaging system for skin cancer diagnosis assistance. In Proceedings of the 2019 XXXIV Conference on Design of Circuits and Integrated Systems (DCIS), Bilbao, Spain, 20–22 November 2019; pp. 1–6.
- Jain, S.; Pise, N. Computer aided melanoma skin cancer detection using image processing. Procedia Comput. Sci. 2015, 48, 735–740.
- Serao, N.; Delfino, K.; Southey, B.; Zas, S.R. Development of a transcriptomic-based index to prognosticate cancer. ISBRA 2010 2010, 2010, 42.
- Carli, P.; Quercioli, E.; Sestini, S.; Stante, M.; Ricci, L.; Brunasso, G.; De Giorgi, V. Pattern analysis, not simplified algorithms, is the most reliable method for teaching dermoscopy for melanoma diagnosis to residents in dermatology. Br. J. Dermatol. 2003, 148, 981–984.
- Kasmi, R.; Mokrani, K. Classification of malignant melanoma and benign skin lesions: Implementation of automatic ABCD rule. IET Image Process. 2016, 10, 448–455.
- Ali, A.-R.H.; Li, J.; Yang, G. Automating the ABCD rule for melanoma detection: A survey. IEEE Access 2020, 8, 83333–83346.
- Ahnlide, I.; Bjellerup, M.; Nilsson, F.; Nielsen, K. Validity of ABCD rule of dermoscopy in clinical practice. Acta Derm. Venereol. 2016, 96, 367–372.
- Binder, M.; Kittler, H.; Steiner, A.; Dawid, M.; Pehamberger, H.; Wolff, K. Reevaluation of the ABCD rule for epiluminescence microscopy. J. Am. Acad. Dermatol. 1999, 40, 171–176.
- Milton, M.A.A. Automated skin lesion classification using ensemble of deep neural networks in ISIC 2018: Skin lesion analysis towards melanoma detection challenge. arXiv 2019, arXiv:1901.10802.
- Basov, S.; Dankner, Y.; Weinstein, M.; Katzir, A.; Platkov, M. Noninvasive mid-IR fiber-optic evanescent wave spectroscopy (FEWS) for early detection of skin cancers. Med. Phys. 2020, 47, 5523–5530.
- Saba, T. Recent advancement in cancer detection using machine learning: Systematic survey of decades, comparisons and challenges. J. Infect. Public Health 2020, 13, 1274–1289.
- Jerant, A.F.; Johnson, J.T.; Sheridan, C.D.; Caffrey, T.J. Early detection and treatment of skin cancer. Am. Fam. Physician 2000, 62, 357–368.
- Ragab, M.; Choudhry, H.; Al-Rabia, M.W.; Binyamin, S.S.; Aldarmahi, A.A.; Mansour, R.F. Early and accurate detection of melanoma skin cancer using hybrid level set approach. Front. Physiol. 2022, 13, 965630.
- Masood, A.; Ali Al-Jumaily, A. Computer aided diagnostic support system for skin cancer: A review of techniques and algorithms. Int. J. Biomed. Imaging 2013, 2013, 323268.
- Johr, R.H. Dermoscopy: Alternative melanocytic algorithms—The ABCD rule of dermatoscopy, menzies scoring method, and 7-point checklist. Clin. Dermatol. 2002, 20, 240–247.
- Kumar, Y.; Gupta, S.; Singla, R.; Hu, Y.-C. A systematic review of artificial intelligence techniques in cancer prediction and diagnosis. Arch. Comput. Methods Eng. 2021, 29, 2043–2070.
- Abbasi, N.R.; Shaw, H.M.; Rigel, D.S.; Friedman, R.J.; McCarthy, W.H.; Osman, I.; Kopf, A.W.; Polsky, D. Early diagnosis of cutaneous melanoma: Revisiting the ABCD criteria. JAMA 2004, 292, 2771–2776.
- Burlina, P.; Billings, S.; Joshi, N.; Albayda, J. Automated diagnosis of myositis from muscle ultrasound: Exploring the use of machine learning and deep learning methods. PLoS ONE 2017, 12, e0184059.
- Chilamkurthy, S.; Ghosh, R.; Tanamala, S.; Biviji, M.; Campeau, N.G.; Venugopal, V.K.; Mahajan, V.; Rao, P.; Warier, P. Deep learning algorithms for detection of critical findings in head CT scans: A retrospective study. Lancet 2018, 392, 2388–2396.
- Bianconi, F.; Fravolini, M.L.; Pizzoli, S.; Palumbo, I.; Minestrini, M.; Rondini, M.; Nuvoli, S.; Spanu, A.; Palumbo, B. Comparative evaluation of conventional and deep learning methods for semi-automated segmentation of pulmonary nodules on CT. Quant. Imaging Med. Surg. 2021, 11, 3286.
- Javed, R.; Rahim, M.S.M.; Saba, T.; Rehman, A. A comparative study of features selection for skin lesion detection from dermoscopic images. Netw. Model. Anal. Health Inform. Bioinform. 2020, 9, 4.
- Hagerty, J.R.; Stanley, R.J.; Almubarak, H.A.; Lama, N.; Kasmi, R.; Guo, P.; Drugge, R.J.; Rabinovitz, H.S.; Oliviero, M.; Stoecker, W.V. Deep learning and handcrafted method fusion: Higher diagnostic accuracy for melanoma dermoscopy images. IEEE J. Biomed. Health Inform. 2019, 23, 1385–1391.
- Zhu, W.; Zeng, N.; Wang, N. Sensitivity, specificity, accuracy, associated confidence interval and ROC analysis with practical SAS implementations. NESUG Proc. Health Care Life Sci. Baltim. Md. 2010, 19, 67.
- Matinfar, M.; Shahidi, S.; Feizi, A. Incidence of nonmelanoma skin cancer in renal transplant recipients: A systematic review and meta-analysis. J. Res. Med. Sci. Off. J. Isfahan Univ. Med. Sci. 2018, 23, 14.
- Gandini, S.; Palli, D.; Spadola, G.; Bendinelli, B.; Cocorocchio, E.; Stanganelli, I.; Miligi, L.; Masala, G.; Caini, S. Anti-hypertensive drugs and skin cancer risk: A review of the literature and meta-analysis. Crit. Rev. Oncol. Hematol. 2018, 122, 1–9.
- Sharon, E.; Snast, I.; Lapidoth, M.; Kaftory, R.; Mimouni, D.; Hodak, E.; Levi, A. Laser treatment for non-melanoma skin cancer: A systematic review and meta-analysis. Am. J. Clin. Dermatol. 2021, 22, 25–38.
- Arafa, A.; Mostafa, A.; Navarini, A.A.; Dong, J.-Y. The association between smoking and risk of skin cancer: A meta-analysis of cohort studies. Cancer Causes Control 2020, 31, 787–794.
- Jiyad, Z.; Olsen, C.; Burke, M.; Isbel, N.; Green, A.C. Azathioprine and risk of skin cancer in organ transplant recipients: Systematic review and meta-analysis. Am. J. Transplant. 2016, 16, 3490–3503.
- Glas, A.S.; Lijmer, J.G.; Prins, M.H.; Bonsel, G.J.; Bossuyt, P.M. The diagnostic odds ratio: A single indicator of test performance. J. Clin. Epidemiol. 2003, 56, 1129–1135.
- Duke, A.A.; Bègue, L.; Bell, R.; Eisenlohr-Moul, T. Revisiting the serotonin–aggression relation in humans: A meta-analysis. Psychol. Bull. 2013, 139, 1148.
- Song, J.; Wan, S.; Piao, S.; Knapp, A.K.; Classen, A.T.; Vicca, S.; Ciais, P.; Hovenden, M.J.; Leuzinger, S.; Beier, C. A meta-analysis of 1119 manipulative experiments on terrestrial carbon-cycling responses to global change. Nat. Ecol. Evol. 2019, 3, 1309–1320.
- Cartiff, B.M.; Duke, R.F.; Greene, J.A. The effect of epistemic cognition interventions on academic achievement: A meta-analysis. J. Educ. Psychol. 2021, 113, 477.
- Greene, J.A.; Cartiff, B.M.; Duke, R.F. A meta-analytic review of the relationship between epistemic cognition and academic achievement. J. Educ. Psychol. 2018, 110, 1084.




