
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jing Chen | -- | 3023 | 2023-12-06 10:50:36 | | | |
| 2 | Peter Tang | + 5 word(s) | 3028 | 2023-12-07 02:35:50 | | | | |
| 3 | Jing Chen | -103 word(s) | 2925 | 2023-12-07 03:05:38 | | | | |
| 4 | Jing Chen | Meta information modification | 2925 | 2023-12-14 16:06:26 | | |
Video Upload Options
Insects harbor diverse assemblages of bacterial and fungal symbionts, which play crucial roles in host life history. Insects and their various symbionts represent a good model for studying host–microbe interactions. Phylosymbiosis is used to describe an eco-evolutionary pattern, providing a new cross-system trend in the research of host-associated microbiota. The phylosymbiosis pattern is characterized by a significant positive correlation between the host phylogeny and microbial community dissimilarities.
1. Introduction of Phylosymbiosis
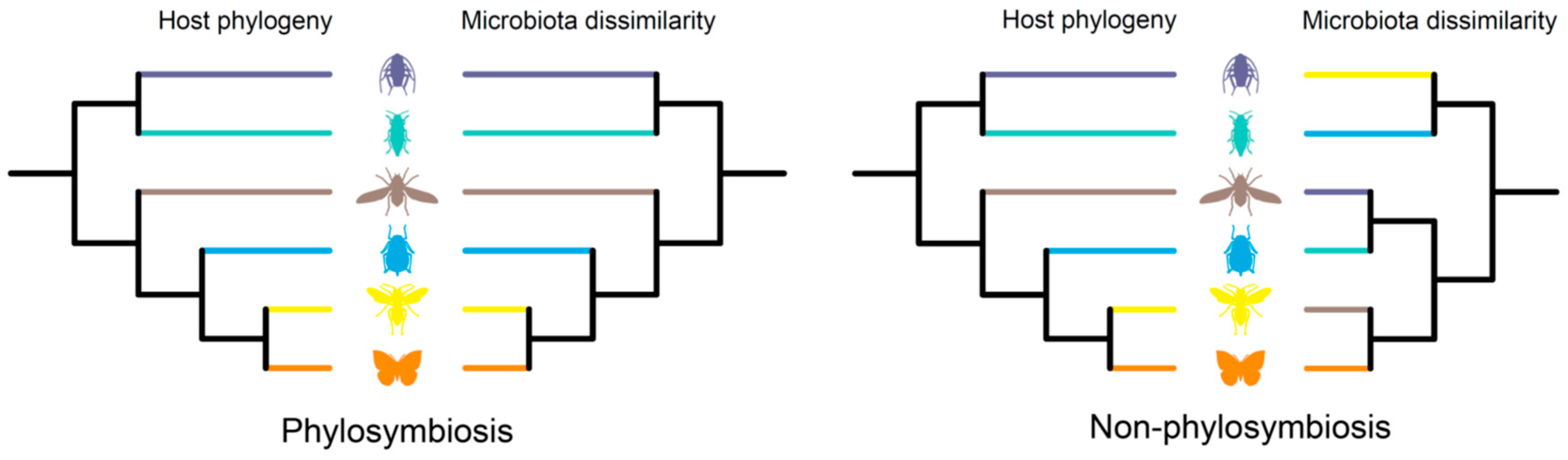
2. Phylosymbiosis in Insects
|
Insects Examined |
No. of Species Sampled |
Evolutionary Time (Mya) |
Diet |
Core Microbe |
Obligate Symbiont |
References |
|
|---|---|---|---|---|---|---|---|
|
Blattodea |
19 |
>300 |
Omnivory |
Bacteroidetes, Firmicutes, and Proteobacteria |
— |
[21] |
|
|
Coleoptera |
Dendroctonus frontalis species complex |
7 |
12 |
Phloem cell |
Ceratocystiopsis |
— |
|
|
Diptera |
Anopheles, Aedes, and Culex |
8 |
100 |
Blood |
Proteobacteria |
— |
|
|
Drosophila |
6 |
63 |
Decaying fruit |
Proteobacteria |
— |
[10] |
|
|
Hemiptera |
Greenideinae |
53 |
83 |
Phloem sap |
— |
Buchnera aphidicola |
|
|
Mollitrichosiphum |
8 |
18–19 |
Phloem sap |
— |
Buchnera aphidicola |
[33] |
|
|
Mollitrichosiphum tenuicorpus |
1 (26 colonies) |
11 |
Phloem sap |
— |
Buchnera aphidicola |
[20] |
|
|
Psylloidea |
102 |
350 |
Phloem sap |
— |
Carsonella ruddii |
||
|
Hymenoptera |
Cephalotes |
13 |
46 |
Pollen and honeydew |
— |
Cephaloticoccus |
[41] |
|
Ceratosolen |
6 |
60 |
Fig |
Wolbachia |
— |
||
|
Formica |
14 |
30 |
Honeydew and nectar |
Wolbachia, Lactobacillus, Liliensternia, and Spiroplasma |
|||
|
Nasonia |
4 |
<1 |
Fly puparium |
Proteobacteria, Firmicutes, and Actinobacteria |
— |
[10] |
|
|
Lepidoptera |
Heliconiini |
23 |
20–30 |
Pollen, nectar, and fruit |
Acinetobacter, Apibacter, Asaia, Commensalibacter, Enterobacter, Enterococcus, Lactococcus, Spiroplasma, and Pseudomonas |
— |
|
3. Mechanisms Underlying Phylosymbiosis
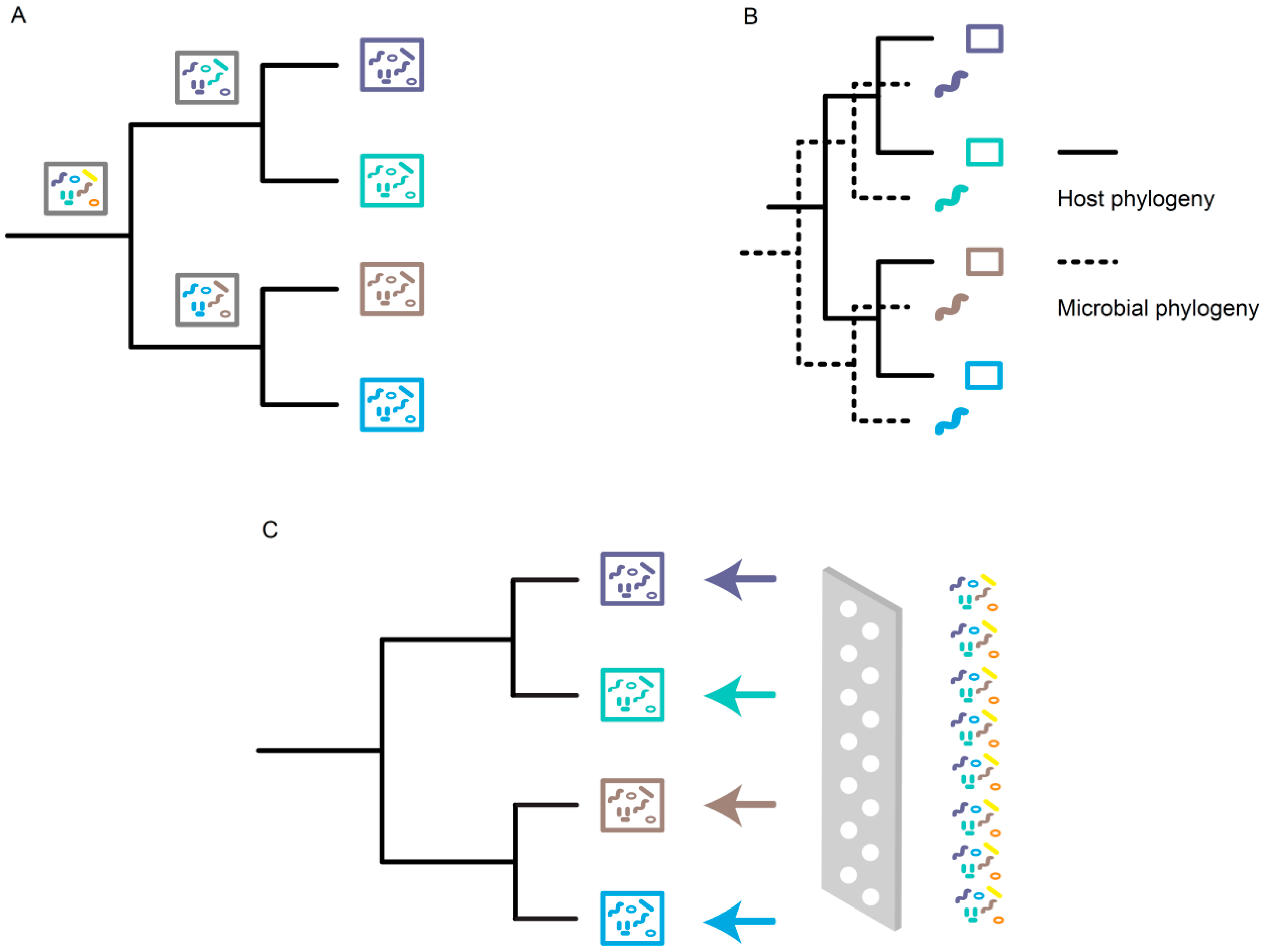
3.1. Stochastic Effects
3.2. Evolutionary Processes
3.3. Ecological Filtering
If the ecological factors that shape microbiota structures are highly phylogenetically conserved during host evolutionary history, we can observe a phylosymbiotic relationship between the host and microbiota [8]. Here, the researchers provide several potential ecological factors shaping the phylosymbiotic microbiota of insects.
3.3.1. Immune System
Numerous studies have highlighted the importance of the host immune system in regulating microbial community composition [95][96][97][98]. Insects rely on physiological barriers and innate immune responses to defend themselves against pathogens [99][100]. The innate immune system of insects is composed of cellular immune responses by circulating hemocytes [101] and humoral immune responses. Although the hemocyte categories involved in cellular immune responses vary among different insect species, hemocyte functions primarily include phagocytosis, nodulation, and encapsulation [102][103][104]. Humoral defenses are modulated by the Toll, immune deficiency (IMD), Jun N-terminal kinase (JNK), Janus kinase/signal transducers and activators of transcription (JAK/STAT), and prophenoloxidase (PPO) pathways [100][105]. The expression of genes in these pathways subsequently results in antimicrobial peptide (AMP) production, reactive oxygen species (ROS) generation, and melanization. Insects depend on two pathways to regulate antimicrobial peptide generation, namely, the Toll pathway, which responds to fungi and most Gram-positive bacteria, and the IMD pathway, which is induced by Gram-negative bacteria [106].
The insect innate immune system not only defends against pathogens but also plays an important role in maintaining host–microbe symbiosis [107][108][109]. Serving as one of the model systems in Hemiptera, aphids lack several immune-related genes that are suspected to be essential in arthropod immunity [110]. Previous studies suggested that the reduced antimicrobial defense in aphid immunity is attributed to the maintenance of symbionts [111]. To be more specific, the extent of alteration in multiple aphid cellular immunity responses is related to the difference in facultative symbiont species [112]. Eusocial corbiculate bees, including honey bees, bumblebees, and stingless bees, harbor distinctive gut microbiota that are more similar among closely related bee species [78]. The exotic strain of the gut symbiont Gilliamella in honey bees induced higher prostaglandin (PG) production than the native strain, which increased the expression of genes in the IMD and Toll immune pathways [113]. These immune pathways then modulated the dual oxidase (Duox) production and ROS generation to inhibit the nonnative strain of Gilliamella.
3.3.2. Diet
There is increasing evidence that diet plays pivotal roles in shaping the microbiota structures of animals [16][32][114][115]. Diet has emerged as a key filter of mammalian gut microbiota [116][117]. The gut microbiota of nonflying mammals was strongly correlated with diet and host phylogeny [115]. Likewise, the microbial communities in bamboo-feeding insects were filtered by diet [31]. If diets themselves are phylogenetically nonindependent, they can serve as ecological filters and lead to phylosymbiotic microbiota. Moreover, complete dietary shifts over long evolutionary periods can disrupt the phylosymbiotic relationship between host and microbial communities [5].
Host plants are one of the major ecological factors shaping the bacterial communities of insect herbivores [31][32][118][119]. The gut microbial communities of caterpillars are dominated by transient and diet-associated bacteria [120], whereas major members of the adult-stage gut microbiota in butterflies are abundant and consistent [46]. The phylosymbiotic signature of microbiota within heliconiine butterflies may arise from the filtering of phylogenetically conserved diet preferences [46]. Within aphids, host–symbiont codiversification as well as filtering by host plants has been highlighted in structuring the phylosymbiotic microbiota of Greenideinae species [37].
3.3.3. Physiological Characteristics
Another candidate ecological filter underlying host species-specific microbiota is host physiological structure, such as gut [121] and proventriculus [122]. Biomolecules such as glycans and mucins secreted by the host intestinal wall shape different intestinal environments and are regulators of gut microbial community composition [123][124]. Other host-specific physical and chemical factors in the gut, including biochemical characteristics of the intestinal surface, pH, oxygen levels, and concentrations of metabolites, are also potential filtering factors of microbes. If these factors themselves are phylogenetically conserved over evolutionary history, the microbial communities might exhibit significant correlations with host phylogeny.
The selective filtering of microbes in the gut environment can explain major variations in the phylosymbiotic gut microbial communities in humans [55]. Compared with mammals, birds (e.g., cranes) have strong gastric acidity, which can serve as a microbial filter to limit host-associated differentiation in gut microbiota and subsequently result in weak phylosymbiotic signatures [14]. In insects, selective effects of the gut environment were experimentally confirmed in the cockroach gut microbiota [125]. Cockroaches preferentially select bacteria that are specifically adapted to their intestinal environment. The proventricular filtering mechanism in ants is responsible for the maintenance of ant–bacteria fidelity [122]. Although the importance of host physiological characteristics in filtering gut microbiota has been emphasized in certain insect groups, its role in shaping insect phylosymbiosis remains poorly understood.
4. Future Directions for Research on Phylosymbiosis in Insects
While host–symbiont interactions have been documented across many insect groups, we still have a poor understanding of the prevalence of phylosymbiosis in insects. Phylosymbiotic investigations should be performed on a greater variety of insects to sufficiently disentangle the mechanisms underlying this pattern. In addition to bacterial and fungal communities, phylosymbiosis studies at the insect–virome level [126][127] will contribute to developing a comprehensive landscape of host–microbe symbioses. The application of metagenomic sequencing data in phylosymbiosis detection is recommended due to its finer-scale taxonomic and functional profiling. Integrated multi-omic analyses of the microbiome are advantageous in comprehending the mechanisms behind phylosymbiosis because they resolve linkages between host functions, microbial diversity, microbial functions, and environmental variables [128].
To date, most studies have focused on the impact of evolutionary processes on driving phylosymbiotic microbiota. Quantifying the contribution of ecological filtering factors in phylosymbiosis will greatly improve the understanding of the mechanisms behind these patterns. Host-specific biological characteristics and environmental factors should be identified and evaluated quantitatively in the future. It is more likely that a combination of multiple mechanisms rather than a single evolutionary or ecological process is involved in the development of phylosymbiosis; therefore, candidate mechanisms, including stochastic effects, evolutionary processes, and ecological filtering, need to be tested in a diversity of symbiosis systems in both evolutionary and ecological contexts.
References
- Perreau, J.; Moran, N.A. Genetic innovations in animal–microbe symbioses. Nat. Rev. Genet. 2022, 23, 23–39.
- Brownlie, J.C.; Johnson, K.N. Symbiont-mediated protection in insect hosts. Trends Microbiol. 2009, 17, 348–354.
- Weinstein, S.B.; Martinez-Mota, R.; Stapleton, T.E.; Klure, D.M.; Greenhalgh, R.; Orr, T.J.; Dale, C.; Kohl, K.D.; Dearing, M.D. Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl. Acad. Sci. USA 2021, 118, e2108787118.
- Youngblut, N.D.; Reischer, G.H.; Walters, W.; Schuster, N.; Walzer, C.; Stalder, G.; Ley, R.E.; Farnleitner, A.H. Host diet and evolutionary history explain different aspects of gut microbiome diversity among vertebrate clades. Nat. Commun. 2019, 10, 2200.
- Groussin, M.; Mazel, F.; Sanders, J.G.; Smillie, C.S.; Lavergne, S.; Thuiller, W.; Alm, E.J. Unraveling the processes shaping mammalian gut microbiomes over evolutionary time. Nat. Commun. 2017, 8, 14319.
- Lim, S.J.; Bordenstein, S.R. An introduction to phylosymbiosis. Proc. R. Soc. B Biol. Sci. 2020, 287, 20192900.
- Brucker, R.M.; Bordenstein, S.R. The hologenomic basis of speciation: Gut bacteria cause hybrid lethality in the genus Nasonia. Science 2013, 341, 667–669.
- Mazel, F.; Davis, K.M.; Loudon, A.; Kwong, W.K.; Groussin, M.; Parfrey, L.W. Is host filtering the main driver of phylosymbiosis across the tree of life? mSystems 2018, 3, e00097-18.
- Brucker, R.M.; Bordenstein, S.R. The roles of host evolutionary relationships (genus: Nasonia) and development in structuring microbial communities. Evolution 2012, 66, 349–362.
- Brooks, A.W.; Kohl, K.D.; Brucker, R.M.; van Opstal, E.J.; Bordenstein, S.R. Phylosymbiosis: Relationships and functional effects of microbial communities across host evolutionary history. PLoS Biol. 2016, 14, e2000225.
- Tang, Y.Y.; Ma, K.Y.; Cheung, M.K.; Yang, C.H.; Wang, Y.Q.; Hu, X.L.; Kwan, H.S.; Chu, K.H. Gut microbiota in decapod shrimps: Evidence of phylosymbiosis. Microb. Ecol. 2021, 82, 994–1007.
- Ding, J.; Jiang, T.; Zhou, H.; Yang, L.; He, C.; Xu, K.; Akinyemi, F.T.; Han, C.; Luo, H.; Qin, C.; et al. The gut microbiota of pheasant lineages reflects their host genetic variation. Front. Genet. 2020, 11, 859.
- Doane, M.P.; Morris, M.M.; Papudeshi, B.; Allen, L.; Pandez, D.; Haggerty, J.M.; Johri, S.; Turnlund, A.C.; Peterson, M.; Kacev, D.; et al. The skin microbiome of elasmobranchs follows phylosymbiosis, but in teleost fishes, the microbiomes converge. Microbiome 2020, 8, 93.
- Trevelline, B.K.; Sosa, J.; Hartup, B.K.; Kohl, K.D. A bird’s-eye view of phylosymbiosis: Weak signatures of phylosymbiosis among all 15 species of cranes. Proc. R. Soc. B Biol. Sci. 2020, 287, 20192988.
- Laviad-Shitrit, S.; Izhaki, I.; Lalzar, M.; Halpern, M. Comparative analysis of intestine microbiota of four wild waterbird species. Front. Microbiol. 2019, 10, 1911.
- Chiarello, M.; Auguet, J.C.; Bettarel, Y.; Bouvier, C.; Claverie, T.; Graham, N.A.J.; Rieuvilleneuve, F.; Sucre, E.; Bouvier, T.; Villeger, S. Skin microbiome of coral reef fish is highly variable and driven by host phylogeny and diet. Microbiome 2018, 6, 147.
- Nishida, A.H.; Ochman, H. Rates of gut microbiome divergence in mammals. Mol. Ecol. 2018, 27, 1884–1897.
- Sanders, J.G.; Powell, S.; Kronauer, D.J.C.; Vasconcelos, H.L.; Frederickson, M.E.; Pierce, N.E. Stability and phylogenetic correlation in gut microbiota: Lessons from ants and apes. Mol. Ecol. 2014, 23, 1268–1283.
- Kohl, K.D.; Varner, J.; Wilkening, J.L.; Dearing, M.D. Gut microbial communities of American pikas (Ochotona princeps): Evidence for phylosymbiosis and adaptations to novel diets. J. Anim. Ecol. 2018, 87, 323–330.
- Qin, M.; Jiang, L.Y.; Kholmatov, B.R.; Chen, J.; Qiao, G.X. Phylosymbiotic structures of the microbiota in Mollitrichosiphum tenuicorpus (Hemiptera: Aphididae: Greenideinae). Microb. Ecol. 2022, 84, 227–239.
- Tinker, K.A.; Ottesen, E.A. Phylosymbiosis across deeply diverging lineages of omnivorous cockroaches (order Blattodea). Appl. Environ. Microbiol. 2020, 86, e02513-19.
- Robinson, D.F.; Foulds, L.R. Comparison of phylogenetic trees. Math. Biosci. 1981, 53, 131–147.
- Bogdanowicz, D.; Giaro, K. On a matching distance between rooted phylogenetic trees. Int. J. Appl. Math. Comput. Sci. 2013, 23, 669–684.
- Mantel, N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967, 27, 209–220.
- Peres-Neto, P.R.; Jackson, D.A. How well do multivariate data sets match? The advantages of a Procrustean superimposition approach over the Mantel test. Oecologia 2001, 129, 169–178.
- Basset, Y.; Cizek, L.; Cuénoud, P.; Didham, R.K.; Guilhaumon, F.; Missa, O.; Novotny, V.; Ødegaard, F.; Roslin, T.; Schmidl, J.; et al. Arthropod diversity in a tropical forest. Science 2012, 338, 1481–1484.
- Perlmutter, J.I.; Bordenstein, S.R. Microorganisms in the reproductive tissues of arthropods. Nat. Rev. Microbiol. 2020, 18, 97–111.
- Douglas, A.E. Mycetocyte symbiosis in insects. Biol. Rev. 1989, 64, 409–434.
- Sudakaran, S.; Kost, C.; Kaltenpoth, M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017, 25, 375–390.
- Malacrinò, A. Host species identity shapes the diversity and structure of insect microbiota. Mol. Ecol. 2022, 31, 723–735.
- Huang, K.G.; Wang, J.; Huang, J.H.; Zhang, S.K.; Vogler, A.P.; Liu, Q.Q.; Li, Y.C.; Yang, M.W.; Li, Y.; Zhou, X.G. Host phylogeny and diet shape gut microbial communities within bamboo–feeding insects. Front. Microbiol. 2021, 12, 633075.
- Yun, J.H.; Roh, S.W.; Whon, T.W.; Jung, M.J.; Kim, M.S.; Park, D.S.; Yoon, C.; Nam, Y.D.; Kim, Y.J.; Choi, J.H.; et al. Insect gut bacterial diversity determined by environmental habitat, diet, developmental stage, and phylogeny of host. Appl. Environ. Microbiol. 2014, 80, 5254–5264.
- Qin, M.; Chen, J.; Xu, S.F.; Jiang, L.Y.; Qiao, G.X. Microbiota associated with Mollitrichosiphum aphids (Hemiptera: Aphididae: Greenideinae): Diversity, host species specificity and phylosymbiosis. Environ. Microbiol. 2021, 23, 2184–2198.
- Vazquez-Ortiz, K.; Pineda-Mendoza, R.M.; González-Escobedo, R.; Davis, T.S.; Salazar, K.F.; Rivera-Orduña, F.N.; Zúñiga, G. Metabarcoding of mycetangia from the Dendroctonus frontalis species complex (Curculionidae: Scolytinae) reveals diverse and functionally redundant fungal assemblages. Front. Microbiol. 2022, 13, 969230.
- Havill, N.P.; Cognato, A.I.; Del-Val, E.K.; Rabaglia, R.J.; Garrick, R.C. New molecular tools for Dendroctonus frontalis (Coleoptera: Curculionidae: Scolytinae) reveal an east-west genetic subdivision of early Pleistocene origin. Insect Syst. Divers. 2019, 3, 817–830.
- Novakova, E.; Woodhams, D.C.; Rodríguez-Ruano, S.M.; Brucker, R.M.; Leff, J.W.; Maharaj, A.; Amir, A.; Knight, R.; Scott, J. Mosquito microbiome dynamics, a background for prevalence and seasonality of West Nile virus. Front. Microbiol. 2017, 8, 526.
- Qin, M.; Chen, J.; Jiang, L.Y.; Qiao, G.X. Insights into species-specific microbiota from Greenideinae (Hemiptera: Aphididae) with evidence of phylosymbiosis. Front. Microbiol. 2022, 13, 828170.
- Liu, Q.H.; Chen, J.; Huang, X.L.; Jiang, L.Y.; Qiao, G.X. Ancient association with Fagaceae in the aphid tribe Greenideini (Hemiptera: Aphididae: Greenideinae). Syst. Entomol. 2015, 40, 230–241.
- Martoni, F.; Bulman, S.R.; Piper, A.M.; Pitman, A.; Taylor, G.S.; Armstrong, K.F. Insect phylogeny structures the bacterial communities in the microbiome of psyllids (Hemiptera: Psylloidea) in Aotearoa New Zealand. PLoS ONE 2023, 18, e0285587.
- Johnson, K.P.; Dietrich, C.H.; Friedrich, F.; Beutel, R.G.; Wipfler, B.; Peters, R.S.; Allen, J.M.; Petersen, M.; Donath, A.; Walden, K.K.O. Phylogenomics and the evolution of hemipteroid insects. Proc. Natl. Acad. Sci. USA 2018, 115, 12775–12780.
- Hu, Y.; D’Amelio, C.L.; Béchade, B.; Cabuslay, C.S.; Łukasik, P.; Sanders, J.G.; Price, S.; Fanwick, E.; Powell, S.; Moreau, C.S.; et al. Partner fidelity and environmental filtering preserve stage-specific turtle ant gut symbioses for over 40 million years. Ecol. Monogr. 2023, 93, e1560.
- Li, J.; Wei, X.; Huang, D.W.; Xiao, J. The phylosymbiosis pattern between the fig wasps of the same genus and their associated microbiota. Front. Microbiol. 2022, 12, 800190.
- Rønsted, N.; Weiblen, G.D.; Cook, J.M.; Salamin, N.; Machado, C.A.; Savolainen, V. 60 million years of co-divergence in the fig-wasp symbiosis. Proc. R. Soc. B Biol. Sci. 2005, 272, 2593–2599.
- Jackson, R.; Patapiou, P.A.; Golding, G.; Helanterä, H.; Economou, C.K.; Chapuisat, M.; Henry, L.M. Evidence of phylosymbiosis in Formica ants. Front. Microbiol. 2023, 14, 1044286.
- Borowiec, M.L.; Cover, S.P.; Rabeling, C. The evolution of social parasitism in Formica ants revealed by a global phylogeny. Proc. Natl. Acad. Sci. USA 2021, 118, e2026029118.
- Hammer, T.J.; Dickerson, J.C.; McMillan, W.O.; Fierer, N. Heliconius butterflies host characteristic and phylogenetically structured adult-stage microbiomes. Appl. Environ. Microbiol. 2020, 86, e02007-20.
- Cicconardi, F.; Milanetti, E.; Pinheiro de Castro, E.C.; Mazo-Vargas, A.; Van Belleghem, S.M.; Ruggieri, A.A.; Rastas, P.; Hanly, J.; Evans, E.; Jiggins, C.D.; et al. Evolutionary dynamics of genome size and content during the adaptive radiation of Heliconiini butterflies. Nat. Commun. 2023, 14, 5620.
- Mallott, E.K.; Amato, K.R. Host specificity of the gut microbiome. Nat. Rev. Microbiol. 2021, 19, 639–653.
- Tung, J.; Barreiro, L.B.; Burns, M.B.; Grenier, J.C.; Lynch, J.; Grieneisen, L.E.; Altmann, J.; Alberts, S.C.; Blekhman, R.; Archie, E.A. Social networks predict gut microbiome composition in wild baboons. eLife 2015, 4, e05224.
- Xu, T.T.; Jiang, L.Y.; Chen, J.; Qiao, G.X. Host plants influence the symbiont diversity of Eriosomatinae (Hemiptera: Aphididae). Insects 2020, 11, 217.
- Xu, T.T.; Chen, J.; Jiang, L.Y.; Qiao, G.X. Diversity of bacteria associated with Hormaphidinae aphids (Hemiptera: Aphididae). Insect Sci. 2021, 28, 165–179.
- Diamond, J.M. Assembly of species communities. In Ecology and Evolution of Communities; Cody, M., Diamond, J.M., Eds.; Belknap Press Harvard University Press: Cambridge, MA, USA, 1975; pp. 342–444.
- Cornwell, W.K.; Schwilk, D.W.; Ackerly, D.D. A trait-based test for habitat filtering: Convex hull volume. Ecology 2006, 87, 1465–1471.
- Weiher, E.; Clarke, G.D.P.; Keddy, P.A. Community assembly rules, morphological dispersion, and the coexistence of plant species. Oikos 1998, 81, 309–322.
- Levy, R.; Borenstein, E. Metabolic modeling of species interaction in the human microbiome elucidates community-level assembly rules. Proc. Natl. Acad. Sci. USA 2013, 110, 12804–12809.
- Kohl, K.D. Ecological and evolutionary mechanisms underlying patterns of phylosymbiosis in host-associated microbial communities. Philos. Trans. R. Soc. B 2020, 375, 20190251.
- Hanson, C.A.; Fuhrman, J.A.; Horner-Devine, M.C.; Martiny, J.B. Beyond biogeographic patterns: Processes shaping the microbial landscape. Nat. Rev. Microbiol. 2012, 10, 497–506.
- Moeller, A.H.; Suzuki, T.A.; Lin, D.; Lacey, E.A.; Wasser, S.K.; Nachman, M.W. Dispersal limitation promotes the diversification of the mammalian gut microbiota. Proc. Natl. Acad. Sci. USA 2017, 114, 13768–13773.
- Nemergut, D.R.; Schmidt, S.K.; Fukami, T.; O’Neill, S.P.; Bilinski, T.M.; Stanish, L.F.; Knelman, J.E.; Darcy, J.L.; Lynch, R.C.; Wickey, P.; et al. Patterns and processes of microbial community assembly. Microbiol. Mol. Biol. Rev. 2013, 77, 342–356.
- Salem, H.; Florez, L.; Gerardo, N.; Kaltenpoth, M. An out-of-body experience: The extracellular dimension for the transmission of mutualistic bacteria in insects. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142957.
- Chase, J.M.; Myers, J.A. Disentangling the importance of ecological niches from stochastic processes across scales. Philos. Trans. R. Soc. B. 2011, 366, 2351–2363.
- Engel, P.; Moran, N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013, 37, 699–735.
- Janzen, D.H. When is it coevolution? Evolution 1980, 34, 611–612.
- Hoberg, E.P.; Brooks, D.R.; Siegel-Causey, D. Host–parasite co-speciation: History, principles, and prospects. In Host–Parasite Evolution: General Principles and Avian Models; Clayton, D.H., Moore, J., Eds.; Oxford University Press: New York, NY, USA, 1997; pp. 212–235.
- Wilson, A.C.; Duncan, R.P. Signatures of host/symbiont genome coevolution in insect nutritional endosymbioses. Proc. Natl. Acad. Sci. USA 2015, 112, 10255–10261.
- Moran, N.A.; McCutcheon, J.P.; Nakabachi, A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008, 42, 165–190.
- Douglas, A.E. How multi-partner endosymbioses function. Nat. Rev. Microbiol. 2016, 14, 731–743.
- Sandström, J.; Moran, N.A. How nutritionally imbalanced is phloem sap for aphids? Entomol. Exp. Appl. 1999, 91, 203–210.
- Manzano-Marín, A.; D’Acier, A.C.; Clamens, A.L.; Orvain, C.; Cruaud, C.; Barbe, V.; Jousselin, E. Serial horizontal transfer of vitamin-biosynthetic genes enables the establishment of new nutritional symbionts in aphids’ di-symbiotic systems. ISME J. 2020, 14, 259–273.
- Luan, J.B.; Chen, W.B.; Hasegawa, D.K.; Simmons, A.M.; Wintermantel, W.M.; Ling, K.S.; Fei, Z.J.; Liu, S.S.; Douglas, A.E. Metabolic coevolution in the bacterial symbiosis of whiteflies and related plant sap-feeding insects. Genome Biol. Evol. 2015, 7, 2635–2647.
- Russell, C.W.; Bouvaine, S.; Newell, P.D.; Douglas, A.E. Shared metabolic pathways in a coevolved insect–bacterial symbiosis. Appl. Environ. Microbiol. 2013, 79, 6117–6123.
- Chong, R.A.; Park, H.; Moran, N.A. Genome evolution of the obligate endosymbiont Buchnera aphidicola. Mol. Biol. Evol. 2019, 36, 1481–1489.
- Moran, N.A. The coevolution of bacterial endosymbionts and phloem-feeding insects. Ann. Mo. Bot. Gard. 2001, 88, 35–44.
- Moran, N.A.; Munson, M.A.; Baumann, P.; Ishikawa, H. A molecular clock in endosymbiotic bacteria is calibrated using the insect hosts. Proc. R. Soc. B Biol. Sci. 1993, 253, 167–171.
- Hosokawa, T.; Kikuchi, Y.; Nikoh, N.; Shimada, M.; Fukatsu, T. Strict host–symbiont cospeciation and reductive genome evolution in insect gut bacteria. PLoS. Biol. 2006, 4, e337.
- Kikuchi, Y.; Hosokawa, T.; Nikoh, N.; Meng, X.Y.; Kamagata, Y.; Fukatsu, T. Host-symbiont co-speciation and reductive genome evolution in gut symbiotic bacteria of acanthosomatid stinkbugs. BMC Biol. 2009, 7, 2.
- Moran, N.A.; Sloan, D.B. The hologenome concept: Helpful or hollow? PLoS Biol. 2015, 13, e1002311.
- Kwong, W.K.; Medina, L.A.; Koch, H.; Sing, K.W.; Soh, E.J.Y.; Ascher, J.S.; Jaffé, R.; Moran, N.A. Dynamic microbiome evolution in social bees. Sci. Adv. 2017, 3, e1600513.
- De Vienne, D.M.; Refrégier, G.; López-Villavicencio, M.; Tellier, A.; Hood, M.E.; Giraud, T.J.N.P. Cospeciation vs host-shift speciation: Methods for testing, evidence from natural associations and relation to coevolution. New Phytol. 2013, 198, 347–385.
- Wiley, E.O. Vicariance biogeography. Annu. Rev. Ecol. Syst. 1988, 19, 513–542.
- Fukami, T. Historical contingency in community assembly: Integrating niches, species pools, and priority effects. Annu. Rev. Ecol. Evol. Syst. 2015, 46, 1–23.
- Debray, R.; Herbert, R.A.; Jaffe, A.L.; Crits-Christoph, A.; Power, M.E.; Koskella, B. Priority effects in microbiome assembly. Nat. Rev. Microbiol. 2022, 20, 109–121.
- Herren, C.M.; McMahon, K.D. Keystone taxa predict compositional change in microbial communities. Environ. Microbiol. 2018, 20, 2207–2217.
- Agler, M.T.; Ruhe, J.; Kroll, S.; Morhenn, C.; Kim, S.; Weigel, D.; Kemen, E. Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 2016, 14, e1002352.
- Fisher, C.K.; Mehta, P. Identifying keystone species in the human gut microbiome from metagenomic tieseries using sparse linear regression. PLoS ONE 2014, 9, e102451.
- Baumann, P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu. Rev. Microbiol. 2005, 59, 155–189.
- Koga, R.; Meng, X.Y.; Tsuchida, T.; Fukatsu, T. Cellular mechanism for selective vertical transmission of an obligate insect symbiont at the bacteriocyte-embryo interface. Proc. Natl. Acad. Sci. USA 2012, 109, E1230–E1237.
- Watanabe, K.; Yukuhiro, F.; Matsuura, Y.; Fukatsu, T.; Noda, H. Intrasperm vertical symbiont transmission. Proc. Natl. Acad. Sci. USA 2014, 111, 7433–7437.
- Kikuchi, Y.; Ohbayashi, T.; Jang, S.; Mergaert, P. Burkholderia insecticola triggers midgut closure in the bean bug Riptortus pedestris to prevent secondary bacterial infections of midgut crypts. ISME J. 2020, 14, 1627–1638.
- Marsh, S.E.; Poulsen, M.; Pinto-Tomas, A.; Currie, C.R. Interaction between workers during a short time window is required for bacterial symbiont transmission in Acromyrmex leaf-cutting ants. PLoS ONE 2014, 9, e103269.
- Powell, J.E.; Martinson, V.G.; Urban-Mead, K.; Moran, N.A. Routes of acquisition of the gut microbiota of the honey bee Apis mellifera. Appl. Environ. Microbiol. 2014, 80, 7378–7387.
- Smith, W.P.J.; Wucher, B.R.; Nadell, C.D.; Foster, K.R. Bacterial defences: Mechanisms, evolution and antimicrobial resistance. Nat. Rev. Microbiol. 2023, 21, 519–534.
- Mason, C.J.; Raffa, K.F. Acquisition and structuring of midgut bacterial communities in gypsy moth (Lepidoptera: Erebidae) larvae. Environ. Entomol. 2014, 43, 595–604.
- Franzenburg, S.; Walter, J.; Künzel, S.; Wang, J.; Baines, J.F.; Bosch, T.C.; Fraune, S. Distinct antimicrobial peptide expression determines host species-specific bacterial associations. Proc. Natl. Acad. Sci. USA 2013, 110, E3730–E3738.
- Kwong, W.K.; Mancenido, A.L.; Moran, N.A. Immune system stimulation by the native gut microbiota of honey bees. R. Soc. Open Sci. 2017, 4, 170003.
- Guarneri, A.A.; Schaub, G.A. Interaction of triatomines, trypanosomes and microbiota. In Triatominae—The Biology of Chagas Disease Vectors; Guarneri, A.A., Lorenzo, M.G., Eds.; Springer Nature: New York, NY, USA, 2021; pp. 345–386.
- Mistry, R.; Kounatidis, I.; Ligoxygakis, P. Interaction between familial transmission and a constitutively active immune system shapes gut microbiota in Drosophila melanogaster. Genetics 2017, 206, 889–904.
- Hooper, L.V.; Littman, D.R.; Macpherson, A.J. Interactions between the microbiota and the immune system. Science 2012, 336, 1268–1273.
- Chen, K.; Lu, Z. Immune responses to bacterial and fungal infections in the silkworm, Bombyx mori. Dev. Comp. Immunol. 2018, 83, 3–11.
- Lemaitre, B.; Hoffmann, J. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 2007, 25, 697–743.
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14.
- Arteaga, B.L.A.; Crispim, J.S.; Fernandes, K.M.; de Oliveira, L.L.; Pereira, M.F.; Bazzolli, D.M.S.; Martins, G.F. Differential cellular immune response of Galleria mellonella to Actinobacillus pleuropneumoniae. Cell Tissue Res. 2017, 370, 153–168.
- Wu, G.Q.; Liu, Y.; Ding, Y.; Yi, Y.H. Ultrastructural and functional characterization of circulating hemocytes from Galleria mellonella larva: Cell types and their role in the innate immunity. Tissue Cell 2016, 48, 297–304.
- Azambuja, P.D.; Garcia, E.S.; Ratcliffe, N.A. Aspects of classification of Hemiptera hemocytes from six triatomine species. Mem. Inst. Oswaldo Cruz. 1991, 86, 1–10.
- Evans, J.D.; Aronstein, K.; Chen, Y.P.; Hetru, C.; Imler, J.L.; Jiang, H.; Kanost, M.; Thompson, G.J.; Zou, Z.; Hultmark, D. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol. Biol. 2006, 15, 645–656.
- Ali Mohammadie Kojour, M.; Han, Y.S.; Jo, Y.H. An overview of insect innate immunity. Entomol. Res. 2020, 50, 282–291.
- Nichols, H.L.; Goldstein, E.B.; Saleh Ziabari, O.; Parker, B.J. Intraspecific variation in immune gene expression and heritable symbiont density. PLoS Pathog. 2021, 17, e1009552.
- Pan, X.; Pike, A.; Joshi, D.; Bian, G.; McFadden, M.J.; Lu, P.; Liang, X.; Zhang, F.; Raikhel, A.S.; Xi, Z. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. 2018, 12, 277–288.
- Nyholm, S.V.; Graf, J. Knowing your friends: Invertebrate innate immunity fosters beneficial bacterial symbioses. Nat. Rev. Microbiol. 2012, 10, 815–827.
- Gerardo, N.M.; Altincicek, B.; Anselme, C.; Atamian, H.; Barribeau, S.M.; de Vos, M.; Duncan, E.J.; Evans, J.D.; Gabaldon, T.; Ghanim, M.; et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010, 11, R21.
- Altincicek, B.; Gross, J.; Vilcinskas, A. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum. Insect Mol. Biol. 2008, 17, 711–716.
- Laughton, A.M.; Garcia, J.R.; Gerardo, N.M. Condition-dependent alteration of cellular immunity by secondary symbionts in the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 2016, 86, 17–24.
- Guo, L.; Tang, J.; Tang, M.; Luo, S.; Zhou, X. Reactive oxygen species are regulated by immune deficiency and Toll pathways in determining the host specificity of honeybee gut bacteria. Proc. Natl. Acad. Sci. USA 2023, 120, e2219634120.
- Alessandri, G.; Rizzo, S.M.; Ossiprandi, M.C.; van Sinderen, D.; Ventura, M. Creating an atlas to visualize the biodiversity of the mammalian gut microbiota. Curr. Opin. Biotechnol. 2022, 73, 28–33.
- Song, S.J.; Sanders, J.G.; Delsuc, F.; Metcalf, J.; Amato, K.; Taylor, M.W.; Mazel, F.; Lutz, H.L.; Winker, K.; Graves, G.R.; et al. Comparative analyses of vertebrate gut microbiomes reveal convergence between birds and bats. mBio 2020, 11, e02901-19.
- Hicks, A.L.; Lee, K.J.; Couto-Rodriguez, M.; Patel, J.; Sinha, R.; Guo, C.; Olson, S.H.; Seimon, A.; Seimon, T.A.; Ondzie, A.U.; et al. Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat. Commun. 2018, 9, 1786.
- Muegge, B.D.; Kuczynski, J.; Knights, D.; Clemente, J.C.; Gonzalez, A.; Fontana, L.; Henrissat, B.; Knight, R.; Gordon, J.I. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science 2011, 332, 970–974.
- Xu, S.F.; Jiang, L.Y.; Qiao, G.X.; Chen, J. The bacterial flora associated with the polyphagous aphid Aphis gossypii Glover (Hemiptera: Aphididae) is strongly affected by host plants. Microb. Ecol. 2020, 79, 971–984.
- Brady, C.M.; Asplen, M.K.; Desneux, N.; Heimpel, G.E.; Hopper, K.R.; Linnen, C.R.; Oliver, K.M.; Wulff, J.A.; White, J.A. Worldwide populations of the aphid Aphis craccivora are infected with diverse facultative bacterial symbionts. Microb. Ecol. 2014, 67, 195–204.
- Hammer, T.J.; Janzen, D.H.; Hallwachs, W.; Jaffe, S.P.; Fierer, N. Caterpillars lack a resident gut microbiome. Proc. Natl. Acad. Sci. USA 2017, 114, 9641–9646.
- Amato, K.R.; Sanders, J.G.; Song, S.J.; Nute, M.; Metcalf, J.L.; Thompson, L.R.; Morton, J.T.; Amir, A.J.; McKenzie, V.; Humphrey, G. Evolutionary trends in host physiology outweigh dietary niche in structuring primate gut microbiomes. ISME J. 2019, 13, 576–587.
- Lanan, M.C.; Rodrigues, P.A.P.; Agellon, A.; Jansma, P.; Wheeler, D.E. A bacterial filter protects and structures the gut microbiome of an insect. ISME J. 2016, 10, 1866–1876.
- McLoughlin, K.; Schluter, J.; Rakoff-Nahoum, S.; Smith, A.L.; Foster, K.R. Host selection of microbiota via differential adhesion. Cell Host Microbe 2016, 19, 550–559.
- Hooper, L.V.; Gordon, J.I. Glycans as legislators of host–microbial interactions: Spanning the spectrum from symbiosis to pathogenicity. Glycobiology 2001, 11, 1R–10R.
- Mikaelyan, A.; Thompson, C.L.; Hofer, M.J.; Brune, A. Deterministic assembly of complex bacterial communities in guts of germ-free cockroaches. Appl. Environ. Microbiol. 2016, 82, 1256–1263.
- Xu, Y.; Jiang, J.Y.; Lin, X.J.; Shi, W.P.; Cao, C. Identification of diverse viruses associated with grasshoppers unveils the parallel relationship between host phylogeny and virome composition. Virus Evol. 2022, 8, veac057.
- Leigh, B.A.; Bordenstein, S.R.; Brooks, A.W.; Mikaelyan, A.; Bordenstein, S.R. Finer-scale phylosymbiosis: Insights from insect viromes. mSystems 2018, 3, e00131-18.
- Jiang, D.; Armour, C.R.; Hu, C.; Mei, M.; Tian, C.; Sharpton, T.J.; Jiang, Y. Microbiome multi-omics network analysis: Statistical considerations, limitations, and opportunities. Front. Genet. 2019, 10, 995.





