Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Silvia Di Agostino | -- | 2615 | 2023-11-24 09:49:29 | | | |
| 2 | Sirius Huang | Meta information modification | 2615 | 2023-11-27 02:22:51 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Rago, V.; Perri, A.; Di Agostino, S. Patient-Derived Preclinical Prostate Cancer Models. Encyclopedia. Available online: https://encyclopedia.pub/entry/52025 (accessed on 08 March 2026).
Rago V, Perri A, Di Agostino S. Patient-Derived Preclinical Prostate Cancer Models. Encyclopedia. Available at: https://encyclopedia.pub/entry/52025. Accessed March 08, 2026.
Rago, Vittoria, Anna Perri, Silvia Di Agostino. "Patient-Derived Preclinical Prostate Cancer Models" Encyclopedia, https://encyclopedia.pub/entry/52025 (accessed March 08, 2026).
Rago, V., Perri, A., & Di Agostino, S. (2023, November 24). Patient-Derived Preclinical Prostate Cancer Models. In Encyclopedia. https://encyclopedia.pub/entry/52025
Rago, Vittoria, et al. "Patient-Derived Preclinical Prostate Cancer Models." Encyclopedia. Web. 24 November, 2023.
Copy Citation
To understand the molecular mechanisms of cancer progression, acquired drug resistance, and the metastatic process, the use of preclinical in vitro models that faithfully summarize the properties of the tumor in patients is still a necessity. The tumor is represented by a diverse group of cell clones, and to reproduce in vitro preclinical tumor models, monolayer cell cultures have been supplanted by patient-derived xenograft (PDX) models and cultured organoids derived from the patient (PDO). These models have proved indispensable for the study of the tumor microenvironment (TME) and its interaction with tumor cells.
prostate cancer
patient-derived organoid (PDO)
patient-derived xenograft (PDX)
precision medicine
targeted therapy
1. Introduction
Prostate cancer (PCa) is the most commonly diagnosed malignancy and the second leading cause of death in males [1][2]. Risk factors for this condition include older age, family history, obesity, hypertension, sedentary lifestyle, high testosterone levels, and ethnicity [3]. There are generally no initial or early symptoms, but late symptoms may include renal failure from ureteral obstruction, anemia, bone pain, and paralysis from spinal metastases.
The primary diagnosis is made by prostate-specific antigen (PSA) testing, although the use of this test for screening remains controversial [4]. On the other hand, ultrasound-guided transrectal prostate tissue biopsies (TRUS) are significant [5]. The latest diagnostic tests include the prostate cancer gene3 (PCA3) urine test, measurement of free and total PSA levels, prostate health index (PHI) score, 4K score/multiparametric magnetic resonance imaging (4K/MRI test), exosome test, genomic analysis, magnetic resonance imaging (MRI), the prostate imaging–reporting and data system (PIRADS), and prostate biopsy using MRI fusion techniques [6]. When cancer is localized, it is considered curable; when it spreads outside the prostate, treatment approaches include hormone treatment, chemotherapy, radiopharmaceuticals, immunotherapy, pain medications, bisphosphonates, rank ligand inhibitors, focused radiation, and other targeted therapies [7]. The effects of these therapies depend on the age of the patient, any associated health problems, the histology of the tumor, and the extent of the disease [8].
PCa treatment usually involves androgen deprivation (ADT), surgical therapy, and pharmaceutical management [1][2]. Androgen receptor signaling plays a key role in the development and proliferation of PCa; for this reason, medical castration by androgen deprivation therapy (ADT) represents a mainstay in standard-of-care PCa. However, long-term ADT, in many patients, determines forms of castration-resistant PCa (CRPC), which is manifested by an increase in the levels of prostate-specific antigen despite castration [9]. Despite different treatment options for CRPC, such as hormone therapy, immunotherapy, chemotherapy, radionuclides, and the use of targeted therapies such as PARP inhibitors, the metastatic CRPC (mCRPC) remains a lethal disease with a very low survival rate [9]. The latest PCa treatments are based on the knowledge of predictive molecular signatures to design an optimal targeted therapeutic protocol [10]. To support these experimental therapies, recent studies have indicated PCa-related genetic abnormalities associated with CRPC (i.e., AR, TP53, PTEN, and BRCA 1/2) [10].
2. Patient-Derived Preclinical Models
Different preclinical PCa models have been developed to identify the molecular mechanisms underlying cancer progression and treatment resistance. Three PCa cell lines, namely LNCaP, DU-145, and PC3, are commonly used for in vitro studies [11], but although these cells allow identifying predictors of treatment response and resistance, they do not represent the diversity and the heterogeneity of human prostate cancer. Therefore, in order to identify new therapeutic targets and make progress in oncology, it is necessary to deepen the cellular and functional biology of the tumor. The need has therefore developed to study cancer no longer on 2D cell lines but in patient-derived models that can faithfully summarize and capture the complexity and heterogeneity of the tumor ecosystem. The creation of omics research groups that facilitate the study and analysis of individual patient tumors are crucial.
A patient-derived preclinical model, also known as the patient-derived xenograft (PDX) model, is a research tool used in cancer studies. It involves transplanting tumor tissue directly from a patient into an immunodeficient mouse or other animal models. This allows the tumor to grow and develop in the animal, creating a model that closely mimics the characteristics of the original patient’s tumor [12]. PDX represents a useful preclinical model for overcoming limitations associated with the use of cancer cell lines, since it retains the key molecular aberrations present in patient tumors that are involved in the tumoral growth. In addition, PDXs have the advantage of partly recreating the complexity of the tumor microenvironment (TME), providing a more reliable and clinically predictive model for drug response screening.
However, the most important limitation of PDX is the lack of influence of the immune system as the mice are immunodepleted. Indeed, although the development of severe immunodeficient mice improved the PDX take rate, these models have a limitation of application because the metastatic behavior of cancer cells in severe immunodeficient models differs from the clinical situation. However, the development of “humanized NSGs” showing functional human T cells, NK cells, and monocytes is helping to better understand the role of cancer immunology and the drug response in the context of immune sufficiency [13]. Furthermore, the application of chimeric grafts with neonatal mouse mesenchyme has significantly increased the xenograft survival rate and doubled the proliferation index of xenografted cancer cells [14]. Most importantly, neuroendocrine prostate cancer (NEPC) PDX models have facilitated the interpretation of NEPC biology and drug sensitivities, allowing the matching of the molecular characteristics of subgroups of NEPC with specific targeted therapeutics. In addition, these preclinical models exhibit a significant role in investigating neuroendocrine transdifferentiation, which has been recognized as a therapeutic resistance mechanism [15]. However, despite this encouraging progress, it is crucial to have an extensive collection of NEPC PDX models covering a range of heterogeneities.
Patient-derived organoids (PDO) are three-dimensional cultures of cells derived from patient tumor samples. They are generated by isolating and culturing tumor cells or tissue fragments in a specialized culture system that supports the growth and self-organization of cells into structures that resemble miniature organs or tissues [16]. In particular, the organoids have provided exciting new opportunities for translational research in oncology, enabling the development of patient-representative biobanks that enable testing for personalized therapies and predict responses prior to their use on the patient.
Organoid cultures from CRPC biopsy samples exhibit highly variable growth rates, presumably depending on the heterogeneity of phenotypes observed in patients; in addition, the long time required for CRPC organoids to expand limited their use to test therapeutic responsiveness. Alternatively, the availability of large CRPC organoid cohorts could be useful in identifying classes of responders and non-responders, as well as potential predictive biomarkers. Organoid cultures can be obtained also from PDX, whose success rate is higher than from biopsy (~50%). Cultures of CRPC PDX-derived organoids are models exhibiting genetic and phenotypic heterogeneity, and therefore, results are particularly useful for mechanistic studies, biomarker identification, and evaluation of drug responses [16]. Furthermore, the co-culture of organoids with immune cells isolated from autologous tumor tissues or peripheral blood of healthy donors represents an attractive approach to exploring the interaction between the immune system and tumors and to predicting the response of patients to immunotherapy [17].
A scheme of the workflow for preparing all these patient cell models is summarized in Figure 1.
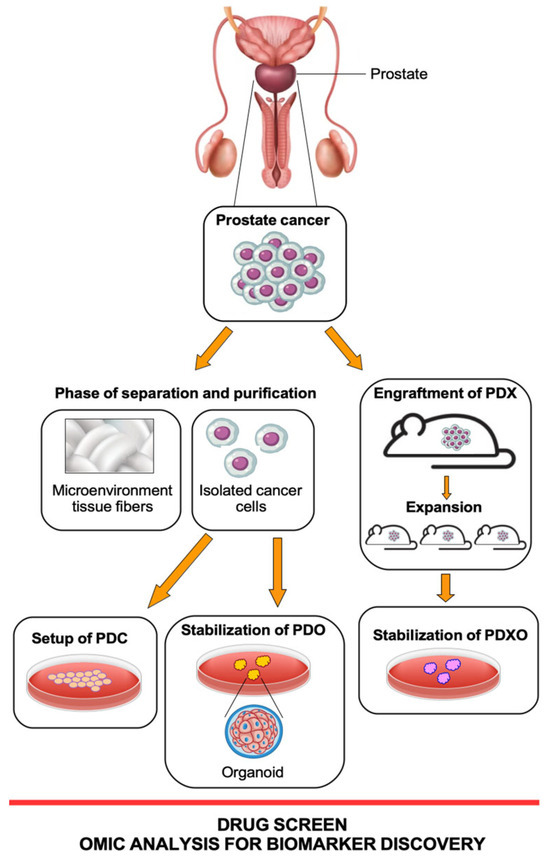
Figure 1. Schematic workflow of three patient-derived models: PDC (patient-derived cells), PDX (patient-derived xenografts), and PDO (patient-derived organoids). PDX-derived organoids (PDXO) are 3D in vitro models generated from patient tumor tissue that have been previously transferred into mouse models for expansion. All these models are derived from tumor tissue from the same PCa patient but throughout different approaches. Figure created at https://www.freepik.com/ (accessed on 1 August 2023).
2.1. Prostate Cancer PDX Models
PDXs are preclinical models that involve transplanting tumor tissue from prostate cancer patients into immunodeficient mice. PCa PDX models capture the heterogeneity of the disease, reflecting the genetic and molecular diversity reported in patients [18][19]. These models are useful in cancer research to study the characteristics of prostate cancer, test potential treatments, and develop personalized therapeutic strategies [20][21].
The duration of tumor settlement in mice can take from a few weeks to a few months to observe the tumor nodule (F0: first generation). After serial transplantation (F1, F2, Fn), the duration of tumor growth becomes stable, and the days needed to obtain tumors of a discrete size depends on the aggressiveness of the PCa [22][23]. Usually, F2/F3 PDX tumors are suitable for cancer biology studies, such as drug sensitivity screening and biomarker identification. The success rate of PDX stabilization varies according to the origin of the tumor and the characteristics of the disease; in PCa, the rate is 65–70% [22][23]. Although the advantage of PDX is to maintain a tumor microenvironment, the loss of human tumor stroma is already observed in the second generation with complete replacement by mouse stroma [24]. The mouse xenograft models that have been used for years are based on immunocompromised mice to avoid rejection of the tissue or injected human tumor cells. Therefore, these mice were found to be inadequate to study the TME in PDXs. To create models versatile in stabilizing a human PDX tumor model, genetically humanized mice and immunologically humanized mice have been developed [13][25].
At the preclinical level, PDXs can be used to identify and validate biomarkers associated with prostate cancer. By comparing the genetic and molecular characteristics of PDX tumors with patient data, several studies reported prediction of treatment response, disease progression, or prognosis [26][27]. Currently, PDX models are the most clinically interesting and adherent in vivo cancer models of drug response obtained from cancer patients [18][28][29]. Thus, the NCI (National Cancer Institute of US) had the initiative to replace a panel of 60 human cell lines (NCI-60) with PDXs for drug testing and the screening of molecules for therapeutic purposes [30].
However, current PCa PDXs, and more generally PDX models of all tumor types, cannot be considered perfectly fitting “avatars” of human cancer. Although PDX in humanized mice for several types of tumors including PCa have been developed, human stromal components of implanted tumor tissue are rapidly lost and replaced by mouse TME during engraftment [31][32][33]. Recently, considering 1110 PDX samples across 24 cancer types, it was documented that PDXs showed rapid accumulation of chromosomal copy number aberrations (CNAs) during the passages, often because of selection of different preexisting cellular clones. CNA acquisition in PDXs was correlated with the genetic heterogeneity observed in primary implanted tumors. Interestingly, some alterations of the copies acquired during PDX passaging differed from those acquired in patients in the course of the disease [33].
Of notable foresight has been a project initiated in 1996 by a group of researchers at the MD Anderson Cancer Center that involved a group of clinically annotated patient-derived xenografts linked to specific phenotypes reflecting all aspects of prostate cancer [34]. The study morphologically characterized 80 PDXs derived from 47 human prostate cancer donors; 47 PDXs derived from 22 donors were working models and were found to be suitable for the growth as cell lines (MDA-PCa 2a and 2b cells) or PDXs [34]. In detail, these PDXs maintained the phenotypic and molecular features of the original tumor, reacting in an expected way to mouse castration and to targeted molecular treatments. Interestingly, supporting the utility of the PDX model in studying tumor heterogeneity, pairs of PDXs derived from different areas of the same tumor showed significant differences in oncogenic pathways, as verified by genomic and RNA sequencing analyses [34] (Table 1).
Very recently, five new PCa PDX models were characterized, including hormone-naïve, androgen-sensitive, and CRPC primary tumors and prostate cancer with neuroendocrine differentiation (NEPC), which is a rare subtype of PCa, and it can either arise de novo or arise as resistance to therapies [35][36]. This biobank was characterized by a comprehensive genomic analysis leading to the identification of driver alterations in androgen signaling, DNA repair, and PI3K pathways, and it was evaluated for androgen deprivation, PARP inhibitors, and chemotherapy response [35]. Overall, these PDXs recapitulated the molecular subtypes of prostate cancer.
Table 1. Benefits and drawbacks of PDX and PDO human-patient-derived models.
| Models | Benefits | Drawbacks |
|---|---|---|
| Mouse PDX-patient-derived Xenografts [12][18][19][25][26][27][28][29][34] |
Possibility to develop the tumor in a physiological TME | Need for an animal house and high costs for maintaining the mice |
| Possibility to study tumor cell heterogeneity in vivo | Long time for engraftment experiments | |
| Crosstalk between factors of the murine immune system and the tumor | High failure rate in engraftment | |
| Possibility to study the response to therapies with in vivo parameters | ||
| PDO-patient-derived Organoids [20][36][37][38][39] |
Limited costs for the formation and maintenance of organoids | Absence of a physiological TME |
| Formation of the organoids in a few days and possibility of amplification in more avatars in the first passages | After a few passages the organoids change the molecular characteristics of the tumor of origin | |
| Organoid ability to grow on scaffolds and mimic signaling as in physiological TME | ||
| Ability to reproduce the structure of the primary tumor tissue |
All these described results provided a wealth of data and insight into the biological mechanisms that could underpin tumor cellular heterogeneity and function as an important resource for the development of personalized therapy targeting specific molecular markers of prostate cancer.
2.2. Prostate Cancer PDO Models
PCa PDOs are three-dimensional cell cultures that are derived from isolated pluripotent stem cells or organ progenitor cells of patient samples, such as tumor tissues or matched healthy tissues [37][38]. The first 3D cultures of patient-derived prostate epithelial cells in the equivalent of artificial basement membrane (BME) matrigel was obtained in the presence of serum, dihydrotestosterone, and stomal cells [39]. These culture conditions, dependent on the number of stem cells in the starting tissue sample, produced morphological differentiation resulting in spheroid-like acini. The first long-term culture of prostate cancer from biopsy specimens and circulating tumor cells occurred in 2014 when the authors reported the first fully molecularly characterized organoids, which recapitulated the molecular diversity of prostate cancer subtypes, including TMPRSS2-ERG fusion, SPOP mutation, SPINK1 overexpression, and CHD1 loss [37].
PCa PDOs were shown to have some disadvantages, such as low efficiency of establishing organoids (15–20% successful in establishing), lack of a physiological TME and immune system, and contamination of normal cells; however, PCa PDOs provide a valuable tool for studying the biology of prostate cancer and testing potential therapeutic strategies [20][37]. PCa PDOs can be used to investigate the genetic and molecular characteristics of individual patient tumors, including their response to different drugs or treatments, having the potential to capture the genomic and clinical heterogeneity observed in prostate cancer and showing a high correlation with the drug response of primary tumors in patients [20] (Table 1).
Prostate cancer is a highly heterogeneous disease, both at the genetic level and in terms of clinical behavior and response to treatment. This heterogeneity poses challenges in understanding the disease and developing effective treatments. Cultures of organoids from CRPC biopsy specimens demonstrate highly variable rates of formation and proliferation, likely demonstrating the heterogeneity of phenotypes observed in patients. Compared to the efficient establishment and rapid growth of organoids from other cancer types, CRPC organoids need a few weeks or even months to grow, to be characterized and used for functional testing [20][40]. This aspect makes the personalized medicine approach for individual PCa patients very complicated, limiting studies for therapeutic responses. However, once the organoid has been created, studies of associated response mechanisms and potential predictive biomarkers have increased in recent years.
Among the disadvantages of culturing organoids there is a lack of cells that populate them, such as fibroblasts, endothelial cells, and immune cells, among others, thus preventing the system from fully recapitulating the complex TME interactions. However, the 3D structures are composed of fully differentiated basal and luminal cells, rare neuroendocrine cells, and cells with stem cell characteristics [41][42]. The presence of TME is also important to determine the success of a treatment. About 90% of preclinically approved drugs do not have the desired effect in clinical trials, partially because of the use of too-simplified in vitro models and the lack of mimicking the TME in drug efficacy screening [42]. Thus, efforts focused on adding the TME components to organoids to recapitulate the microenvironment of parental tumors are ongoing.
References
- Luining, W.I.; Cysouw, M.C.F.; Meijer, D.; Hendrikse, N.H.; Boellaard, R.; Vis, A.N.; Oprea-Lager, D.E. Targeting PSMA Revolutionizes the Role of Nuclear Medicine in Diagnosis and Treatment of Prostate Cancer. Cancers 2022, 14, 1169.
- Sandhu, S.; Moore, C.M.; Chiong, E.; Beltran, H.; Bristow, R.G.; Williams, S.G. Prostate Cancer. Lancet 2021, 398, 1075–1090.
- Kaiser, A.; Haskins, C.; Siddiqui, M.M.; Hussain, A.; D’Adamo, C. The Evolving Role of Diet in Prostate Cancer Risk and Progression. Curr. Opin. Oncol. 2019, 31, 222–229.
- Sadeghi-Nejad, H.; Simmons, M.; Dakwar, G.; Dogra, V. Controversies in Transrectal Ultrasonography and Prostate Biopsy. Ultrasound Q. 2006, 22, 169–175.
- Harvey, C.J.; Pilcher, J.; Richenberg, J.; Patel, U.; Frauscher, F. Applications of transrectal ultrasound in prostate cancer. Br. J. Radiol. 2012, 85, S3–S17.
- Sivaraman, A.; Bhat, K.R.S. Screening and Detection of Prostate Cancer-Review of Literature and Current Perspective. Indian J. Surg. Oncol. 2017, 8, 160–168.
- Wilt, T.J.; Jones, K.M.; Barry, M.J.; Andriole, G.L.; Culkin, D.; Wheeler, T.; Aronson, W.J.; Brawer, M.K. Follow-up of Prostatectomy versus Observation for Early Prostate Cancer. N. Engl. J. Med. 2017, 377, 132–142.
- Loriot, Y.; Massard, C.; Fizazi, K. Recent developments in treatments targeting castration-resistant prostate cancer bone metastases. Ann. Oncol. 2012, 23, 1085–1094.
- Nuhn, P.; De Bono, J.S.; Fizazi, K.; Freedland, S.J.; Grilli, M.; Kantoff, P.W.; Sonpavde, G.; Sternberg, C.N.; Yegnasubramanian, S.; Antonarakis, E.S. Update on Systemic Prostate Cancer Therapies: Management of Metastatic Castration-Resistant Prostate Cancer in the Era of Precision Oncology. Eur. Urol. 2019, 75, 88–99.
- Mateo, J.; McKay, R.; Abida, W.; Aggarwal, R.; Alumkal, J.; Alva, A.; Feng, F.; Gao, X.; Graff, J.; Hussain, M.; et al. Accelerating Precision Medicine in Metastatic Prostate Cancer. Nat. Cancer 2020, 1, 1041–1053.
- Sobel, R.E.; Sadar, M.D. Cell lines used in prostate cancer research: A compendium of old and new lines—Part 1. J. Urol. 2005, 173, 342–359.
- Risbridger, G.P.; Lawrence, M.G.; Taylor, R.A. PDX: Moving Beyond Drug Screening to Versatile Models for Research Discovery. J. Endocr. Soc. 2020, 4, bvaa132.
- Shultz, L.D.; Brehm, M.A.; Garcia-Martinez, J.V.; Greiner, D.L. Humanized Mice for Immune System Investigation: Progress, Promise and Challenges. Nat. Rev. Immunol. 2012, 12, 786–798.
- Toivanen, R.; Berman, D.M.; Wang, H.; Pedersen, J.; Frydenberg, M.; Meeker, A.K.; Ellem, S.J.; Risbridger, G.P.; Taylor, R.A. Brief Report: A Bioassay to Identify Primary Human Prostate Cancer Repopulating Cells. Stem Cells 2011, 29, 1310–1314.
- Shi, M.; Wang, Y.; Lin, D.; Wang, Y. Patient-derived xenograft models of neuroendocrine prostate cancer. Cancer Lett. 2022, 525, 160–169.
- Beshiri, M.; Agarwal, S.; Yin, J.J.; Kelly, K. Prostate Organoids: Emerging Experimental Tools for Translational Research. J. Clin. Investig. 2023, 133, e169616.
- Dijkstra, K.K.; Cattaneo, C.M.; Weeber, F.; Chalabi, M.; van de Haar, J.; Fanchi, L.F.; Slagter, M.; van der Velden, D.L.; Kaing, S.; Kelderman, S.; et al. Generation of Tumor-Reactive T Cells by Co-culture of Peripheral Blood Lymphocytes and Tumor Organoids. Cell 2018, 174, 1586–1598.e12.
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350.
- Cassidy, J.W.; Caldas, C.; Bruna, A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res. 2015, 75, 2963–2968.
- Beshiri, M.L.; Tice, C.M.; Tran, C.; Nguyen, H.M.; Sowalsky, A.G.; Agarwal, S.; Jansson, K.H.; Yang, Q.; McGowen, K.M.; Yin, J.; et al. A PDX/Organoid Biobank of Advanced Prostate Cancers Captures Genomic and Phenotypic Heterogeneity for Disease Modeling and Therapeutic Screening. Clin. Cancer Res. 2018, 24, 4332–4345.
- Shi, C.; Chen, X.; Tan, D. Development of Patient-Derived Xenograft Models of Prostate Cancer for Maintaining Tumor Heterogeneity. Transl. Androl. Urol. 2019, 8, 519–528.
- Mattie, M.; Christensen, A.; Chang, M.S.; Yeh, W.; Said, S.; Shostak, Y.; Capo, L.; Verlinsky, A.; An, Z.; Joseph, I.; et al. Molecular Characterization of Patient-Derived Human Pancreatic Tumor Xenograft Models for Preclinical and Translational Development of Cancer Therapeutics. Neoplasia 2013, 15, 1138–1150.
- Guo, S.; Gao, S.; Liu, R.; Shen, J.; Shi, X.; Bai, S.; Wang, H.; Zheng, K.; Shao, Z.; Liang, C.; et al. Oncological and Genetic Factors Impacting PDX Model Construction with NSG Mice in Pancreatic Cancer. FASEB J. 2019, 33, 873–884.
- Blomme, A.; Van Simaeys, G.; Doumont, G.; Costanza, B.; Bellier, J.; Otaka, Y.; Sherer, F.; Lovinfosse, P.; Boutry, S.; Palacios, A.P.; et al. Murine Stroma Adopts a Human-like Metabolic Phenotype in the PDX Model of Colorectal Cancer and Liver Metastases. Oncogene 2018, 37, 1237–1250.
- Stripecke, R.; Münz, C.; Schuringa, J.J.; Bissig, K.-D.; Soper, B.; Meeham, T.; Yao, L.-C.; Di Santo, J.P.; Brehm, M.; Rodriguez, E.; et al. Innovations, Challenges, and Minimal Information for Standardization of Humanized Mice. EMBO Mol. Med. 2020, 12, e8662.
- Okada, S.; Vaeteewoottacharn, K.; Kariya, R. Application of Highly Immunocompromised Mice for the Establishment of Patient-Derived Xenograft (PDX) Models. Cells 2019, 8, 889.
- Nguyen, L.C.; Naulaerts, S.; Bruna, A.; Ghislat, G.; Ballester, P.J. Predicting Cancer Drug Response In Vivo by Learning an Optimal Feature Selection of Tumour Molecular Profiles. Biomedicines 2021, 9, 1319.
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-Throughput Screening Using Patient-Derived Tumor Xenografts to Predict Clinical Trial Drug Response. Nat. Med. 2015, 21, 1318–1325.
- Pompili, L.; Porru, M.; Caruso, C.; Biroccio, A.; Leonetti, C. Patient-Derived Xenografts: A Relevant Preclinical Model for Drug Development. J. Exp. Clin. Cancer Res. 2016, 35, 189.
- Ledford, H. US Cancer Institute to Overhaul Tumour Cell Lines. Nature 2016, 530, 391.
- Zhao, Y.; Shuen, T.W.H.; Toh, T.B.; Chan, X.Y.; Liu, M.; Tan, S.Y.; Fan, Y.; Yang, H.; Lyer, S.G.; Bonney, G.K.; et al. Development of a New Patient-Derived Xenograft Humanized Mouse Model to Study Human-Specific Tumour Microenvironment and Immunotherapy. Gut 2018, 67, 1845–1854.
- Buqué, A.; Galluzzi, L. Modeling Tumor Immunology and Immunotherapy in Mice. Trends Cancer 2018, 4, 599–601.
- Ben-David, U.; Ha, G.; Tseng, Y.-Y.; Greenwald, N.F.; Oh, C.; Shih, J.; McFarland, J.M.; Wong, B.; Boehm, J.S.; Beroukhim, R.; et al. Patient-Derived Xenografts Undergo Mouse-Specific Tumor Evolution. Nat. Genet. 2017, 49, 1567–1575.
- Palanisamy, N.; Yang, J.; Shepherd, P.D.A.; Li-Ning-Tapia, E.M.; Labanca, E.; Manyam, G.C.; Ravoori, M.K.; Kundra, V.; Araujo, J.C.; Efstathiou, E.; et al. The MD Anderson Prostate Cancer Patient-Derived Xenograft Series (MDA PCa PDX) Captures the Molecular Landscape of Prostate Cancer and Facilitates Marker-Driven Therapy Development. Clin. Cancer Res. 2020, 26, 4933–4946.
- Béraud, C.; Bidan, N.; Lassalle, M.; Lang, H.; Lindner, V.; Krucker, C.; Masliah-Planchon, J.; Potiron, E.; Lluel, P.; Massfelder, T.; et al. A New Tumorgraft Panel to Accelerate Precision Medicine in Prostate Cancer. Front. Oncol. 2023, 13, 1130048.
- Cacciatore, A.; Albino, D.; Catapano, C.V.; Carbone, G.M. Preclinical Models of Neuroendocrine Prostate Cancer. Curr. Protoc. 2023, 3, e742.
- Gao, D.; Vela, I.; Sboner, A.; Iaquinta, P.J.; Karthaus, W.R.; Gopalan, A.; Dowling, C.; Wanjala, J.N.; Undvall, E.A.; Arora, V.K.; et al. Organoid Cultures Derived from Patients with Advanced Prostate Cancer. Cell 2014, 159, 176–187.
- Drost, J.; Karthaus, W.R.; Gao, D.; Driehuis, E.; Sawyers, C.L.; Chen, Y.; Clevers, H. Organoid Culture Systems for Prostate Epithelial and Cancer Tissue. Nat. Protoc. 2016, 11, 347–358.
- Lang, S.H.; Stark, M.; Collins, A.; Paul, A.B.; Stower, M.J.; Maitland, N.J. Experimental Prostate Epithelial Morphogenesis in Response to Stroma and Three-Dimensional Matrigel Culture. Cell Growth Differ. 2001, 12, 631–640.
- Servant, R.; Garioni, M.; Vlajnic, T.; Blind, M.; Pueschel, H.; Müller, D.C.; Zellweger, T.; Templeton, A.J.; Garofoli, A.; Maletti, S.; et al. Prostate Cancer Patient-Derived Organoids: Detailed Outcome from a Prospective Cohort of 81 Clinical Specimens. J. Pathol. 2021, 254, 543–555.
- Puca, L.; Bareja, R.; Prandi, D.; Shaw, R.; Benelli, M.; Karthaus, W.R.; Hess, J.; Sigouros, M.; Donoghue, A.; Kossai, M.; et al. Patient Derived Organoids to Model Rare Prostate Cancer Phenotypes. Nat. Commun. 2018, 9, 2404.
- Trivedi, P.; Liu, R.; Bi, H.; Xu, C.; Rosenholm, J.M.; Åkerfelt, M. 3D Modeling of Epithelial Tumors-The Synergy between Materials Engineering, 3D Bioprinting, High-Content Imaging, and Nanotechnology. Int. J. Mol. Sci. 2021, 22, 6225.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
451
Revisions:
2 times
(View History)
Update Date:
27 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





