Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ioannis Michalopoulos | -- | 2541 | 2023-11-22 16:55:48 | | | |
| 2 | Fanny Huang | Meta information modification | 2541 | 2023-11-23 07:13:16 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Pagoni, M.; Cava, C.; Sideris, D.C.; Avgeris, M.; Zoumpourlis, V.; Michalopoulos, I.; Drakoulis, N. miRNA-Based Technologies in Cancer Therapy. Encyclopedia. Available online: https://encyclopedia.pub/entry/51936 (accessed on 07 March 2026).
Pagoni M, Cava C, Sideris DC, Avgeris M, Zoumpourlis V, Michalopoulos I, et al. miRNA-Based Technologies in Cancer Therapy. Encyclopedia. Available at: https://encyclopedia.pub/entry/51936. Accessed March 07, 2026.
Pagoni, Maria, Claudia Cava, Diamantis C. Sideris, Margaritis Avgeris, Vassilios Zoumpourlis, Ioannis Michalopoulos, Nikolaos Drakoulis. "miRNA-Based Technologies in Cancer Therapy" Encyclopedia, https://encyclopedia.pub/entry/51936 (accessed March 07, 2026).
Pagoni, M., Cava, C., Sideris, D.C., Avgeris, M., Zoumpourlis, V., Michalopoulos, I., & Drakoulis, N. (2023, November 22). miRNA-Based Technologies in Cancer Therapy. In Encyclopedia. https://encyclopedia.pub/entry/51936
Pagoni, Maria, et al. "miRNA-Based Technologies in Cancer Therapy." Encyclopedia. Web. 22 November, 2023.
Copy Citation
The discovery of therapeutic miRNAs is one of the most exciting challenges for pharmaceutical companies. Since the first miRNA was discovered in 1993, our knowledge of miRNA biology has grown considerably. Many studies have demonstrated that miRNA expression is dysregulated in many diseases, making them appealing tools for novel therapeutic approaches.
miRNA
cancer
1. Introduction
Therapeutic agents are categorized mainly into synthetic small molecules, monoclonal antibodies, or large proteins. Traditional drugs may be insufficient to hit intended therapeutic targets because of the inaccessibility of active sites in the target’s three-dimensional structure. RNA-based therapies may offer an excellent chance to potentially reach any therapeutic-relevant target [1] (see Table 1). Additionally, techniques based on nucleic acid technologies are less laborious towards synthesis procedures. Nevertheless, RNA-based therapies have specificity issues that carry the risk of off-target effects [2][3].
Table 1. Methods available for the inhibition of miRNAs.
| Method of Delivery | Characteristics |
|---|---|
| anti-miRNA ASOs (AMOs) | complementary sequence to endogenous miRNA |
| Modified AMOs with 2′-O-methyl-group (OMe) | Increase binding affinity and nuclease resistance |
| Modified AMOs with 2′ methoxyethyl (MOE) | More stable and specific |
| Modified AMOs with locked nucleic acid (LNA) | High binding affinity and low toxicity |
| miRNA sponges | Inhibit a whole family of associated miRNAs |
| miRNA masking | Gene-specific |
In this context, the therapeutic use of miRNA therapy is receiving attention in clinical trials of almost all human diseases Depending on the expression pattern in pathological conditions there are two main streams of miRNA therapies that include the development of synthetic molecules with an effect on protein expression [4]: miRNA mimics [5][6] and miRNA antagonists [7][8]. The restoration of miRNA functions and the inhibition of overexpressed miRNAs are fundamental for the development of miRNA-based cancer therapeutics [9][10]. This is because, the regulation of specific miRNA alterations through miRNA mimics or antagomirs normalizes the gene regulatory network and the signaling pathways, reversing the phenotypes in cancer cells [9][10][11].
Even though miRNA therapeutic potential is still at the preclinical and early clinical stage, researchers are encouraged to invest in miRNAs, as many miRNAs satisfy the stringent criteria that could make them successful therapeutic agents due to their function as oncogenes or tumor suppressors [12][13][14][15].
2. miRNA Inhibitors
The inhibition of oncogenic miRNAs has been achieved by antisense oligonucleotides (anti-microRNA oligonucleotides—AMOs), also called antagomirs, miRNA sponges, and miRNA-Masking Antisense Oligonucleotides [16][17][18][19]. Table 1 and Figure 1 show the main miRNA therapeutic strategies.
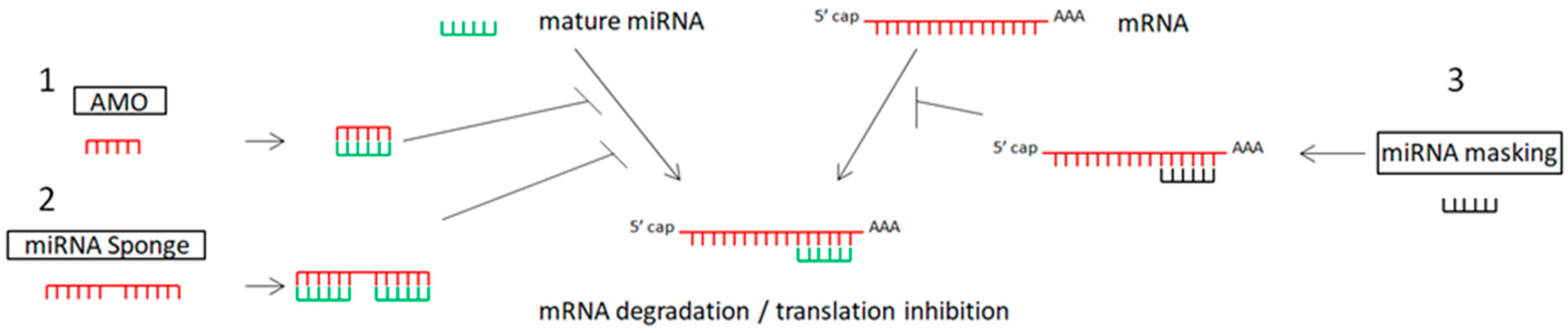
Figure 1. Schematic representation of miRNA therapeutic strategies: 1. anti-miRNA oligonucleotides (AMOs) bind with miRNA, preventing the binding of miRNA to mRNA; 2. miRNA sponges include multiple sites for miRNA binding, blocking the binding of miRNA to mRNA; 3. miRNA mask binds with the specific gene (target of miRNA).
2.1. Anti-miRNA Oligonucleotides (AMOs) or Antagomirs
The most popular approach to inhibit miRNA function is the synthesis of antisense oligonucleotides with complementary sequences to endogenous miRNAs. The chemical structure that characterizes them helps to trap the endogenous miRNA that cannot be further processed by the RISC complex or where the endogenous miRNA is being degraded. During this process, the endogenous miRNA of interest serves as a biomarker optimizing the pharmacokinetic and pharmacodynamic properties of the antagonist [20][21][22][23].
First-generation pre-clinical compounds, the antisense phosphorothiolated oligodeoxynucleotides, were characterized by a low affinity for their target, negative immunostimulatory effects, and they also had very short half-lives, due to their immediate excretion by renal clearance [24][25].
These properties have been improved in the next generation of drug agents with the design of antisense and anti-miR oligonucleotides supported by better pharmacokinetic properties [26]. Because of their capacity to bind cognate sequences, miRNA’s action on target mRNAs can be inhibited by steric antisense oligonucleotides (ASOs) that have high affinity and specificity for some miRNAs [27]. ASOs are single-stranded antisense oligonucleotides, in particular chemically modified DNA-like molecules, 17 to 22 nt in length, which have been designed to have complementarity to a specific mRNA, inhibiting its translation. They have been used for more than thirty years in clinical trials phases II and III [20][28], but they have also been used with success to screen gene functions in high-throughput cellular assays. Their clinical and preclinical application in cancer is for the design of therapeutic solutions together with ribozymes, DNAzymes, small interfering RNAs (siRNAs), short hairpin RNAs (shRNAs), anti-miRNA agents such as ASOs-anti-miRNAs, and locked nucleic acids (LNA)-anti-miRNAs or antagomirs. Although their inhibitory action on the upregulated oncogenic miRNAs has an efficient and long-lasting effect [29], due to their weak pharmacodynamic and pharmacokinetic properties, the delivery of these agents is not ideal [30][31].
Experimental evidence on ASO applications for miR-21 and miR-221 showed increased expression levels of PTEN, RECK, and CDKN1B, but also reduced proliferation and increased apoptosis of pancreatic tumor cells [32][33]. The latest preliminary studies in pancreatic cancer on small side population (SP) cells with stem cell-like properties suggest that inhibition of miR-21 and miR-221 with combinatory ASOs technology against miR-21 and miR-221 inhibited primary tumor growth and metastasis compared to a single antagomir treatment [34]. ASOs can sensitize pancreatic ductal adenocarcinoma cells (PDAC) to gemcitabine causing synergistic anticancer outcomes [33].
A class of antisense oligonucleotides has been subjected to biochemical modifications, and they have been transformed into ssRNA analogs complementary only to specific miRNAs. They are also known under the term anti-miRNA ASOs (AMOs) [35][36]. In this mechanism of action, the modified synthetic anti-miRNA oligonucleotides (AMOs) inhibit specific individual miRNAs through competitive inhibition of base-pairing, as they block the interaction between miRNA and its target mRNAs.
miR-16, miR-21, miR-214, and miR-181a have been identified as potential drug targets for lung cancer therapy by design, synthesis, and benefit-specific AMOs on A549 lung carcinoma cells. Following transfection, some properties, such as cell viability, apoptosis, and miRNA expression, were tested under variable conditions of dose and time. It was observed that AMO-miR-21, AMO-miR-16, and AMO-miR-181a arrested cell growth by inducing apoptosis and S-phase suppression, suggesting that these miRNAs could be potential targets for cancer therapy and AMOs could be a functional technique for miRNA inhibition [37].
Chemical Modifications of AMOs
In the past, several chemical modifications of AMOs have been designed to advance their efficiency and stability such as the addition of 2′-O-methyl and 2′-O-methoxyethyl formations to the 5′ end of the molecule [38].
2′-O-methyl-group (OMe)
The 2′-O-methyl-group (OMe) is one of the most frequent chemical modifications to oligonucleotides. The methyl group has the function to contribute to nuclease resistance improvement and the binding affinity to RNA. Completely modified OMe-oligonucleotides have found application to prevent aberrant exon splicing in cells [39], whereas the hybrid backbone OMe/DNA that is formed in antisense oligonucleotides is under investigation in clinical applications [40]. Synthetic antisense oligonucleotides bearing 2′-O-Methyl (2′OMe) ribose modification and antagomirs were shown to decrease miRISC activity and to inhibit specifically miRNAs that have acquired a gain of function in human diseases including cancer [20][28].
Synthetic AMOs that bring the 2-O-methyl modification showed effective inhibition of target miRNAs in cell and xerograph models with the disadvantage of operating in high doses in the given study [41]. 2-O-methyl AMOs have been applied as inhibitors in glioblastoma and breast cancer in human cell lines and xenografts by targeting miR-21 [42]. Today, 2′OMe-modified AMOs represent the most used molecules to dissect miRNA roles, not only due to their high affinity for targeting miRNAs but also for the lack of high levels of toxicity and economic cost of synthesis [43].
2′-O-Methoxyethyl
2′-O-Methoxyethyl (MOE)-modified oligonucleotides show evidence of higher binding affinity and specificity to RNA compared to OMe-analogs. They have been used efficiently as chemically modified oligonucleotides with the property to re-address mRNA splicing. They also inhibit protein translation. MOE-ASOs are increasingly considered important modified oligonucleotides in clinical trials [44]. The combination of antisense oligonucleotides (ASOs) with 2′-O-(2-methoxyethyl) (2′-MOE) is a platform of RNA-based therapeutics with the property to hybridize to their target RNA via Watson–Crick base pairing, preventing the expression of the “disease-related” protein product. Recently, there has been high interest expressed in the number of 2′-MOE ASOs progressing to phase I, II, and III clinical trials for therapies in many diseases, including rheumatoid arthritis [45], cancer [46], and hypercholesterolemia [47].
Locked nucleic acid (LNA)
The chemistry-locked nucleic acid (LNA)-modified oligonucleotides consist of a 2′-O-modified RNA molecule where the 2′-O-oxygen links to the 4′-position through a methylene linker, making a bicyclic compound locked into a C3′-endo (RNA) sugar [48]. The LNA chemical modification provides thermodynamic stability to the duplex conformation with known RNA molecules. The use of LNA oligonucleotides as northern probes is important for the detection of miRNAs as mixed backbone oligonucleotides [49]. Potentially, they could be a new class of therapy. Among the types of nucleoside modifications applied, the addition of chemical groups to the 2′-hydroxyl group has been quite successful [50]. In addition, oligonucleotide derivatives could be applicable as curative AMOs [51].
Preclinical studies focusing on the understanding of the mechanisms describing osteosarcoma-initiating cells and their potential clinical significance showed that deregulation of miR-133a in a highly malignant CD133 cellular population affects cell invasiveness and identifies a lethal tumor phenotype. The inhibition of this miRNA by LNA reduced cell invasion, whereas administration of LNA in addition to chemotherapy suppressed lung cancer metastasis and increased survival outcomes in osteosarcoma-bearing mice. Clinically, overexpression of both CD133 and miR-133a has been associated with poor prognosis, whereas overexpression of four CD133 targets correlates to a good prognosis. Overall, silencing LNA miR-133a in combination with chemotherapy was proposed as an anticancer strategy, developed at the preclinical stage for targeting multiple regulatory pathways associated with metastasis of osteosarcoma cells [52]. In addition, studies investigating the effectiveness of such inhibitory approaches documented that although 2′ modifications were shown to improve affinity to target RNAs, their anti-miRNA activity was not fully correlated with affinity, suggesting that other variables may also be important for effective miRNA [53]. Studies analyzing the tissue toxicity of LNA in mice and monkeys focused on the effect induced by LNA-miR-221 inhibitors in vital organs. They confirmed the low toxicity of LNA and suggested its use for clinical purposes [54].
2.2. miRNA Sponges in Cancer Therapy
The miRNA “sponge” technology was introduced in 2007 with the purpose of ensuring miRNA loss of function in cell models and transgenic organisms. miRNA sponges are usually plasmid-encoding copies that contain binding sites complementary to the seed region of the target miRNA [7][55] and are products of transgenes within cells.
Following cell transfection, these plasmids transcribe high levels of sponge RNAs that bind to the seed region, which enables them to block a family of miRNAs that contain the same seed sequence. As competitive inhibitors, miRNA sponges exhibit similar inhibition efficiency with short nucleotide fragments [7][55]. In addition, it has been shown that miRNA sponges that are based on retroviral vectors are able to knock down miRNAs but also block an entire miRNA seed family using one vector [56].
The sponge binding sites act on the miRNA seeding region, and, in this way, they block a whole family of related miRNAs. The fine-tuning of miRNA sponge concentration compared to miRNA target concentration is crucial for the efficacy of miRNA inhibition. The highest expression of miRNA sponge is achieved when the affinity and avidity of binding sites are high, but also when the promoter used in the cell model of interest is strong, i.e., a cytomegalovirus promoter. Sponge inhibitors are used in long-term miRNA loss-of-function research and in vivo assays, for example, bone marrow reconstitution and cancer xenografts. The stability of miRNA sponge activity by expressing the transgene from chromosomal integrations has been tested by different groups [57][58][59]. The importance of miRNA sponges in cancer therapy is to mimic the down-regulation of specific miRNAs that are deregulated.
Before investigating the biological effects of miR-21 on A549 non-small cell lung cancer (NSCLC) cells, the expression of miR-21 in serum samples of patients affected by NSCLC was quantified. More precisely, they used miRNA sponge technology and transfection of A549 cells, suggesting that miR-21 could be an independent molecular biomarker for NSCLC, but also that modulating miR-21 or PDCD4 expression may provide a potentially novel therapeutic approach for NSCLC [60].
Competing Endogenous RNAs (ceRNAs)
Competing endogenous RNAs (ceRNAs) act as natural endogenous miRNA sponges, regulating the bioavailability and function of miRNAs. They include transcribed pseudogenes, long non-coding RNAs (lncRNAs), and circular RNAs (circRNAs). Their synergic action forming molecular networks aims to the regulation of protein expression [61]. Their advantage compared to antisense oligonucleotides, which target a single miRNA, is that they can have numerous different binding sites, coordinating simultaneous inhibition of a big subset of a miRNA cluster, or of distinct miRNAs that act on the same target. The development of quantitative methods for the determination of the absolute expression levels of miRNA and ceRNA molecules allows the possibility of estimating the efficiency of ceRNA crosstalk in many biological models. In addition, the use of mathematical models contributes to the calculation of fluctuations in ceRNA and miRNA levels, which respond to variations in additional parameters, such as structure and topology [62].
2.3. miRNA-Masking
The development of miRNA-mask technology that includes miRNA-masking antisense oligonucleotides uses single-stranded 2′-O-methyl-modified antisense oligonucleotides, which are complementary to the miRNA binding sites in the 3′-UTR of the target mRNA [57]. Due to the masking effect of these miRNAs to the target mRNA, their action is gene-specific. Recently, miRNA therapeutics technology used constructs of miRNA-masking oligonucleotide drugs (ONDs) that attach to mRNA. The binding of an miRNA-mask to its target site is assisted by the use of O-methyl groups and also LNAs, to improve the masking efficiency [63]. The high efficiency of miRNA masking is based on target gene selection. miRNA–gene interactions that are crucial for tumorigenesis, such as miRNA-203 and LASP1, and miRNA-29-b-1 and SPIN1, which are involved in proliferation, angiogenesis, migration, and apoptosis, have been investigated [64].
3. miRNA Mimics
miRNA replacement therapy is gaining traction worldwide using synthetic miRNAs or mimics, which, when incorporated, can restore the normal physiological activity of the organism. The miRNA replacement strategy is classified as either a viral or non-viral delivery therapy [65]. The overexpression of miRNAs can also be achieved by the synthesis of miRNA mimics, which are designed for protein-coding gene silencing [6][66]. The biochemistry of miRNA replacement technology includes an active strand of the mimic, which contains a sequence that is normally expressed to the cell, whereas the passenger strand is chemically modified to achieve the interaction of the active strand with the protein complex and guarantee the functionality of the miRNA.
miRNA mimics can also be described as double-stranded-like RNAs, which are composed frequently of siRNA-like oligoribonucleotide duplexes. [67][68]. They have a sequence motif on their 5′ end that is partially complementary to the target sequence in the 3′-UTR of the target mRNA. They mimic the functionality of mature endogenous miRNAs. The use of mimics is important for miRNA functionality assessment because it provides a tool for gain-of-function studies. The restoration of miRNAs that show a loss of function through miRNA mimics is fundamentally used to explore the therapeutic potentiality of tumor suppressors [69][70].
Technically, miRNA replacement therapy acts downstream of the miRISC complex and requires the enzymatic functions of cellular miRISC to be catalytically functional. miRNA mimics target multiple transcripts, and it seems that regulate the same set of genes as the endogenous miRNAs.
It was observed that the overexpression of miR-21 associated with keratinization of tumors in cases of oral squamous cell carcinomas was significantly correlated with the poor prognosis of patients. Transfection of miR-7- and miR-21-mimics reduced the expression of RECK, which is a tumor suppressor gene, through direct miRNA-mediated regulation. This study provided important information related to patient survival, with the aim of contributing to improved therapeutics for oral cancer [71].
References
- Iacomino, G. miRNAs: The Road from Bench to Bedside. Genes 2023, 14, 314.
- Zhu, Y.; Zhu, L.; Wang, X.; Jin, H. RNA-based therapeutics: An overview and prospectus. Cell Death Dis. 2022, 13, 644.
- Jo, S.J.; Chae, S.U.; Lee, C.B.; Bae, S.K. Clinical Pharmacokinetics of Approved RNA Therapeutics. Int. J. Mol. Sci. 2023, 24, 746.
- Das, N.; Tripathi, N.; Khurana, S. Micro RNA Mimics And Antagonists. Int. J. Sci. Technol. Res. 2015, 4, 176–180.
- Manikkath, J.; Jishnu, P.V.; Wich, P.R.; Manikkath, A.; Radhakrishnan, R. Nanoparticulate strategies for the delivery of miRNA mimics and inhibitors in anticancer therapy and its potential utility in oral submucous fibrosis. Nanomedicine 2022, 17, 181–195.
- Kang, E.; Kortylewski, M. Lipid Nanoparticle-Mediated Delivery of miRNA Mimics to Myeloid Cells. Methods Mol. Biol. 2023, 2691, 337–350.
- Fu, Y.; Chen, J.; Huang, Z. Recent progress in microRNA-based delivery systems for the treatment of human disease. ExRNA 2019, 1, 24.
- Reda El Sayed, S.; Cristante, J.; Guyon, L.; Denis, J.; Chabre, O.; Cherradi, N. MicroRNA Therapeutics in Cancer: Current Advances and Challenges. Cancers 2021, 13, 2680.
- He, B.; Zhao, Z.; Cai, Q.; Zhang, Y.; Zhang, P.; Shi, S.; Xie, H.; Peng, X.; Yin, W.; Tao, Y.; et al. miRNA-based biomarkers, therapies, and resistance in Cancer. Int. J. Biol. Sci. 2020, 16, 2628–2647.
- Fu, Z.; Wang, L.; Li, S.; Chen, F.; Au-Yeung, K.K.; Shi, C. MicroRNA as an Important Target for Anticancer Drug Development. Front. Pharmacol. 2021, 12, 736323.
- Otoukesh, B.; Abbasi, M.; Gorgani, H.O.; Farahini, H.; Moghtadaei, M.; Boddouhi, B.; Kaghazian, P.; Hosseinzadeh, S.; Alaee, A. MicroRNAs signatures, bioinformatics analysis of miRNAs, miRNA mimics and antagonists, and miRNA therapeutics in osteosarcoma. Cancer Cell Int. 2020, 20, 254.
- Check Hayden, E. Cancer complexity slows quest for cure. Nature 2008, 455, 148.
- Parsons, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An integrated genomic analysis of human glioblastoma multiforme. Science 2008, 321, 1807–1812.
- Medina, P.P.; Nolde, M.; Slack, F.J. OncomiR addiction in an in vivo model of microRNA-21-induced pre-B-cell lymphoma. Nature 2010, 467, 86–90.
- Menon, A.; Abd-Aziz, N.; Khalid, K.; Poh, C.L.; Naidu, R. miRNA: A Promising Therapeutic Target in Cancer. Int. J. Mol. Sci. 2022, 23, 11502.
- Garzon, R.; Marcucci, G.; Croce, C.M. Targeting microRNAs in cancer: Rationale, strategies and challenges. Nat. Rev. Drug Discov. 2010, 9, 775–789.
- Cannataro, R.; Cione, E. miRNA as Drug: Antagomir and Beyond. Curr. Pharm. Des. 2023, 29, 462–465.
- Ortega, M.M.; Bouamar, H. Guidelines on Designing MicroRNA Sponges: From Construction to Stable Cell Line. Methods Mol. Biol. 2023, 2595, 171–183.
- Takegawa-Araki, T.; Kumagai, S.; Yasukawa, K.; Kuroda, M.; Sasaki, T.; Obika, S. Structure-Activity Relationships of Anti-microRNA Oligonucleotides Containing Cationic Guanidine-Modified Nucleic Acids. J. Med. Chem. 2022, 65, 2139–2148.
- Hutvagner, G.; Simard, M.J.; Mello, C.C.; Zamore, P.D. Sequence-specific inhibition of small RNA function. PLoS Biol. 2004, 2, E98.
- Orom, U.A.; Kauppinen, S.; Lund, A.H. LNA-modified oligonucleotides mediate specific inhibition of microRNA function. Gene 2006, 372, 137–141.
- Zhang, J.; Sharma, R.; Ryu, K.; Shen, P.; Salaita, K.; Jo, H. Conditional Antisense Oligonucleotides Triggered by miRNA. ACS Chem. Biol. 2021, 16, 2255–2267.
- Quemener, A.M.; Bachelot, L.; Forestier, A.; Donnou-Fournet, E.; Gilot, D.; Galibert, M.D. The powerful world of antisense oligonucleotides: From bench to bedside. Wiley Interdiscip. Rev. RNA 2020, 11, e1594.
- Seth, P.P.; Siwkowski, A.; Allerson, C.R.; Vasquez, G.; Lee, S.; Prakash, T.P.; Wancewicz, E.V.; Witchell, D.; Swayze, E.E. Short antisense oligonucleotides with novel 2′-4′ conformationaly restricted nucleoside analogues show improved potency without increased toxicity in animals. J. Med. Chem. 2009, 52, 10–13.
- Shadid, M.; Badawi, M.; Abulrob, A. Antisense oligonucleotides: Absorption, distribution, metabolism, and excretion. Expert. Opin. Drug Metab. Toxicol. 2021, 17, 1281–1292.
- Monia, B.P.; Lesnik, E.A.; Gonzalez, C.; Lima, W.F.; McGee, D.; Guinosso, C.J.; Kawasaki, A.M.; Cook, P.D.; Freier, S.M. Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as antisense inhibitors of gene expression. J. Biol. Chem. 1993, 268, 14514–14522.
- Lennox, K.A.; Behlke, M.A. Chemical modification and design of anti-miRNA oligonucleotides. Gene Ther. 2011, 18, 1111–1120.
- Meister, G.; Landthaler, M.; Dorsett, Y.; Tuschl, T. Sequence-specific inhibition of microRNA- and siRNA-induced RNA silencing. Rna 2004, 10, 544–550.
- Esau, C.C. Inhibition of microRNA with antisense oligonucleotides. Methods 2008, 44, 55–60.
- Spizzo, R.; Rushworth, D.; Guerrero, M.; Calin, G.A. RNA inhibition, microRNAs, and new therapeutic agents for cancer treatment. Clin. Lymphoma Myeloma 2009, 9 (Suppl. S3), S313–S318.
- Zhang, S.; Chen, L.; Jung, E.J.; Calin, G.A. Targeting microRNAs with small molecules: From dream to reality. Clin. Pharmacol. Ther. 2010, 87, 754–758.
- Moriyama, T.; Ohuchida, K.; Mizumoto, K.; Yu, J.; Sato, N.; Nabae, T.; Takahata, S.; Toma, H.; Nagai, E.; Tanaka, M. MicroRNA-21 modulates biological functions of pancreatic cancer cells including their proliferation, invasion, and chemoresistance. Mol. Cancer Ther. 2009, 8, 1067–1074.
- Park, J.K.; Lee, E.J.; Esau, C.; Schmittgen, T.D. Antisense inhibition of microRNA-21 or -221 arrests cell cycle, induces apoptosis, and sensitizes the effects of gemcitabine in pancreatic adenocarcinoma. Pancreas 2009, 38, e190–e199.
- Zhao, Y.; Zhao, L.; Ischenko, I.; Bao, Q.; Schwarz, B.; Niess, H.; Wang, Y.; Renner, A.; Mysliwietz, J.; Jauch, K.W.; et al. Antisense inhibition of microRNA-21 and microRNA-221 in tumor-initiating stem-like cells modulates tumorigenesis, metastasis, and chemotherapy resistance in pancreatic cancer. Target. Oncol. 2015, 10, 535–548.
- Krutzfeldt, J.; Rajewsky, N.; Braich, R.; Rajeev, K.G.; Tuschl, T.; Manoharan, M.; Stoffel, M. Silencing of microRNAs in vivo with ‘antagomirs’. Nature 2005, 438, 685–689.
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352.
- Fei, J.; Lan, F.; Guo, M.; Li, Y.; Liu, Y. Inhibitory effects of anti-miRNA oligonucleotides (AMOs) on A549 cell growth. J. Drug Target. 2008, 16, 688–693.
- Baker, B.F.; Lot, S.S.; Condon, T.P.; Cheng-Flournoy, S.; Lesnik, E.A.; Sasmor, H.M.; Bennett, C.F. 2′-O-(2-Methoxy)ethyl-modified anti-intercellular adhesion molecule 1 (ICAM-1) oligonucleotides selectively increase the ICAM-1 mRNA level and inhibit formation of the ICAM-1 translation initiation complex in human umbilical vein endothelial cells. J. Biol. Chem. 1997, 272, 11994–12000.
- Friedman, K.J.; Kole, J.; Cohn, J.A.; Knowles, M.R.; Silverman, L.M.; Kole, R. Correction of aberrant splicing of the cystic fibrosis transmembrane conductance regulator (CFTR) gene by antisense oligonucleotides. J. Biol. Chem. 1999, 274, 36193–36199.
- Mani, S.; Goel, S.; Nesterova, M.; Martin, R.M.; Grindel, J.M.; Rothenberg, M.L.; Zhang, R.; Tortora, G.; Cho-Chung, Y.S. Clinical studies in patients with solid tumors using a second-generation antisense oligonucleotide (GEM 231) targeted against protein kinase A type I. Ann. N. Y. Acad. Sci. 2003, 1002, 252–262.
- Trang, P.; Medina, P.P.; Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Homer, R.; Brown, D.; Bader, A.G.; Weidhaas, J.B.; et al. Regression of murine lung tumors by the let-7 microRNA. Oncogene 2010, 29, 1580–1587.
- Si, M.L.; Zhu, S.; Wu, H.; Lu, Z.; Wu, F.; Mo, Y.Y. miR-21-mediated tumor growth. Oncogene 2007, 26, 2799–2803.
- Lennox, K.A.; Owczarzy, R.; Thomas, D.M.; Walder, J.A.; Behlke, M.A. Improved Performance of Anti-miRNA Oligonucleotides Using a Novel Non-Nucleotide Modifier. Mol. Ther.-Nucleic Acids 2013, 2, e117.
- Gleave, M.E.; Monia, B.P. Antisense therapy for cancer. Nat. Rev. Cancer 2005, 5, 468–479.
- Sewell, K.L.; Geary, R.S.; Baker, B.F.; Glover, J.M.; Mant, T.G.; Yu, R.Z.; Tami, J.A.; Dorr, F.A. Phase I trial of ISIS 104838, a 2′-methoxyethyl modified antisense oligonucleotide targeting tumor necrosis factor-alpha. J. Pharmacol. Exp. Ther. 2002, 303, 1334–1343.
- Chi, K.N.; Siu, L.L.; Hirte, H.; Hotte, S.J.; Knox, J.; Kollmansberger, C.; Gleave, M.; Guns, E.; Powers, J.; Walsh, W.; et al. A phase I study of OGX-011, a 2′-methoxyethyl phosphorothioate antisense to clusterin, in combination with docetaxel in patients with advanced cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 833–839.
- Raal, F.J.; Santos, R.D.; Blom, D.J.; Marais, A.D.; Charng, M.J.; Cromwell, W.C.; Lachmann, R.H.; Gaudet, D.; Tan, J.L.; Chasan-Taber, S.; et al. Mipomersen, an apolipoprotein B synthesis inhibitor, for lowering of LDL cholesterol concentrations in patients with homozygous familial hypercholesterolaemia: A randomised, double-blind, placebo-controlled trial. Lancet 2010, 375, 998–1006.
- Vester, B.; Wengel, J. LNA (locked nucleic acid): High-affinity targeting of complementary RNA and DNA. Biochemistry 2004, 43, 13233–13241.
- Valoczi, A.; Hornyik, C.; Varga, N.; Burgyan, J.; Kauppinen, S.; Havelda, Z. Sensitive and specific detection of microRNAs by northern blot analysis using LNA-modified oligonucleotide probes. Nucleic Acids Res. 2004, 32, e175.
- Henry, S.P.; Geary, R.S.; Yu, R.; Levin, A.A. Drug properties of second-generation antisense oligonucleotides: How do they measure up to their predecessors? Curr. Opin. Investig. Drugs 2001, 2, 1444–1449.
- Weiler, J.; Hunziker, J.; Hall, J. Anti-miRNA oligonucleotides (AMOs): Ammunition to target miRNAs implicated in human disease? Gene Ther. 2006, 13, 496–502.
- Fujiwara, T.; Katsuda, T.; Hagiwara, K.; Kosaka, N.; Yoshioka, Y.; Takahashi, R.U.; Takeshita, F.; Kubota, D.; Kondo, T.; Ichikawa, H.; et al. Clinical relevance and therapeutic significance of microRNA-133a expression profiles and functions in malignant osteosarcoma-initiating cells. Stem Cells 2014, 32, 959–973.
- Davis, S.; Lollo, B.; Freier, S.; Esau, C. Improved targeting of miRNA with antisense oligonucleotides. Nucleic Acids Res. 2006, 34, 2294–2304.
- Gallo Cantafio, M.E.; Nielsen, B.S.; Mignogna, C.; Arbitrio, M.; Botta, C.; Frandsen, N.M.; Rolfo, C.; Tagliaferri, P.; Tassone, P.; Di Martino, M.T. Pharmacokinetics and Pharmacodynamics of a 13-mer LNA-inhibitor-miR-221 in Mice and Non-human Primates. Mol. Ther. Nucleic Acids 2016, 5, E326.
- Jie, J.; Liu, D.; Wang, Y.; Wu, Q.; Wu, T.; Fang, R. Generation of MiRNA sponge constructs targeting multiple MiRNAs. J. Clin. Lab. Anal. 2022, 36, e24527.
- Ebert, M.S.; Neilson, J.R.; Sharp, P.A. MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian cells. Nat. Methods 2007, 4, 721–726.
- Xiao, J.; Yang, B.; Lin, H.; Lu, Y.; Luo, X.; Wang, Z. Novel approaches for gene-specific interference via manipulating actions of microRNAs: Examination on the pacemaker channel genes HCN2 and HCN4. J. Cell. Physiol. 2007, 212, 285–292.
- Papapetrou, E.P.; Korkola, J.E.; Sadelain, M. A genetic strategy for single and combinatorial analysis of miRNA function in mammalian hematopoietic stem cells. Stem Cells 2010, 28, 287–296.
- Ma, L.; Young, J.; Prabhala, H.; Pan, E.; Mestdagh, P.; Muth, D.; Teruya-Feldstein, J.; Reinhardt, F.; Onder, T.T.; Valastyan, S.; et al. miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol. 2010, 12, 247–256.
- Yang, Y.; Meng, H.; Peng, Q.; Yang, X.; Gan, R.; Zhao, L.; Chen, Z.; Lu, J.; Meng, Q.H. Downregulation of microRNA-21 expression restrains non-small cell lung cancer cell proliferation and migration through upregulation of programmed cell death 4. Cancer Gene Ther. 2015, 22, 23–29.
- Bak, R.O.; Mikkelsen, J.G. miRNA sponges: Soaking up miRNAs for regulation of gene expression. Wiley Interdiscip. Rev. RNA 2014, 5, 317–333.
- Krell, J.; Frampton, A.E.; Jacob, J.; Castellano, L.; Stebbing, J. miRNAs in breast cancer: Ready for real time? Pharmacogenomics 2012, 13, 709–719.
- Murakami, K.; Miyagishi, M. Tiny masking locked nucleic acids effectively bind to mRNA and inhibit binding of microRNAs in relation to thermodynamic stability. Biomed. Rep. 2014, 2, 509–512.
- Colangelo, T.; Polcaro, G.; Ziccardi, P.; Muccillo, L.; Galgani, M.; Pucci, B.; Milone, M.R.; Budillon, A.; Santopaolo, M.; Mazzoccoli, G.; et al. The miR-27a-calreticulin axis affects drug-induced immunogenic cell death in human colorectal cancer cells. Cell Death Dis. 2016, 7, e2108.
- Saiyed, A.N.; Vasavada, A.R.; Johar, S.R.K. Recent trends in miRNA therapeutics and the application of plant miRNA for prevention and treatment of human diseases. Futur. J. Pharm. Sci. 2022, 8, 24.
- Yang, H.; Liu, Y.; Chen, L.; Zhao, J.; Guo, M.; Zhao, X.; Wen, Z.; He, Z.; Chen, C.; Xu, L. MiRNA-Based Therapies for Lung Cancer: Opportunities and Challenges? Biomolecules 2023, 13, 877.
- Hossain, A.; Kuo, M.T.; Saunders, G.F. Mir-17-5p regulates breast cancer cell proliferation by inhibiting translation of AIB1 mRNA. Mol. Cell. Biol. 2006, 26, 8191–8201.
- Hutvagner, G.; Zamore, P.D. RNAi: Nature abhors a double-strand. Curr. Opin. Genet. Dev. 2002, 12, 225–232.
- Takeshita, F.; Patrawala, L.; Osaki, M.; Takahashi, R.U.; Yamamoto, Y.; Kosaka, N.; Kawamata, M.; Kelnar, K.; Bader, A.G.; Brown, D.; et al. Systemic delivery of synthetic microRNA-16 inhibits the growth of metastatic prostate tumors via downregulation of multiple cell-cycle genes. Mol. Ther. J. Am. Soc. Gene Ther. 2010, 18, 181–187.
- Wiggins, J.F.; Ruffino, L.; Kelnar, K.; Omotola, M.; Patrawala, L.; Brown, D.; Bader, A.G. Development of a lung cancer therapeutic based on the tumor suppressor microRNA-34. Cancer Res. 2010, 70, 5923–5930.
- Jung, H.M.; Phillips, B.L.; Patel, R.S.; Cohen, D.M.; Jakymiw, A.; Kong, W.W.; Cheng, J.Q.; Chan, E.K. Keratinization-associated miR-7 and miR-21 regulate tumor suppressor reversion-inducing cysteine-rich protein with kazal motifs (RECK) in oral cancer. J. Biol. Chem. 2012, 287, 29261–29272.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
912
Entry Collection:
Biopharmaceuticals Technology
Revisions:
2 times
(View History)
Update Date:
23 Nov 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





