In the past century, microbial natural products have been a primary source of pharmaceuticals attributing to their structural diversities and fruitful bioactivities
[1]. For example, in 1928, British microbiologist Alexander Fleming accidentally discovered penicillin from microorganisms, and thereafter, the use of penicillin in controlling bacterial infections has made great success. Encouraged by these positive results and the demand for the treatment of other diseases, the discovery of antibiotics has entered a golden age, and peaked in the mid-1950s
[2]. However, one of the greatest medical dilemmas of the 20th century is the presence of bacterial resistance to antibiotics which are rapidly becoming ineffective in clinical practice, a sharp and seemly unsolvable problem most probably originated from the broad judicious and injudicious use of antibiotics, as well as the incorrect disposal of their waste
[3]. In order to solve this problem, several measures have been taken at different levels. These measures are listed as follows: (1) discovering new antibiotics; (2) exploring new biological targets in microbial cells; (3) chemical modification of the existing antibiotics; (4) rationally controlling the use of antibiotics and carefully treating their wastes; (5) developing vaccines. Among all these measures, the discovery of novel bioactive antibiotics by using modern techniques to avoid the rediscovery of secondary metabolites from microbes are promising to alleviate this problem
[4]. Indeed, natural products coming from plants, animals, or microorganisms, and synthetic compounds designed based on natural scaffold play an important role in new drug discovery. For instance, from the 1970s to 2019, one-third of the approved drug breakthroughs were related to natural products, including unaltered natural product (3.8%), natural “Botanical” ones (0.8%), and their semi-synthetic derivatives or analogues (18.9%). In addition, there are many delicate total synthetic drugs whose pharmacophores were also inspired by natural products
[5]. However, the less availability of novel bioactive secondary metabolites and the increasing rate at which known compounds are being rediscovered make this approach increasingly disappointing. Delightedly, on the basis of natural products, the utilization of genomics, synthetic biology, chemical informatics, bioinformatics, or synthetic chemistry has significantly advanced the discovery of drug candidates, which were widely used in a diverse range of human diseases
[5][6].
2. How to Pair Strains for Co-Culture?
2.1. Based on the Ecological Niches
Niche refers to an area that is occupied by different microorganism populations which communicate with each other to form a special ecosystem under certain temporal and spatial latitude conditions. Microorganisms that live and reproduce in this ecological environment compete and cooperate due to limited nutritional resources.
Marine-derived endophytes are found to exist between or within the cells of marine organisms, including marine plants (algae, seagrass, driftwood, and mangrove), marine vertebrates (mainly fish), and marine invertebrates (mainly sponges and corals)
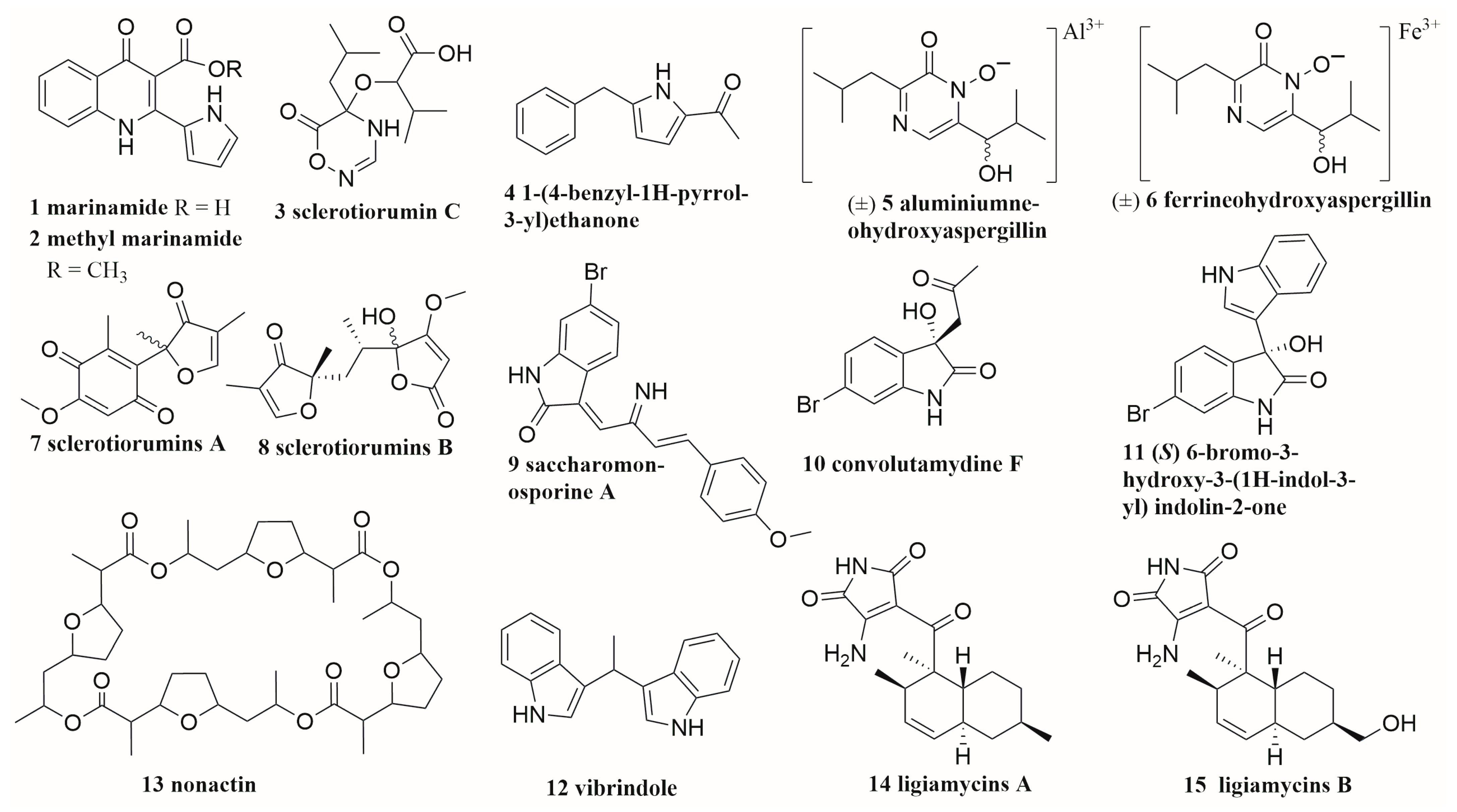
[14]. Zhu et al. co-cultured two unidentified mangrove endophytic fungi (Nos. 1924 and 3893, isolated from the coast of South China Sea) and isolated two new compounds (
1,
2) (
Figure 1)
[15]. Bao et al. isolated six new secondary metabolites (
3–
6,
7,
8) from the co-culture extracts of marine-derived fungi
Aspergillus sclerotiorum and
Penicillium citrinum. The two strains were isolated from gorgonian
Muricella flexuosa collected from the South China Sea
[16]. In 2018, Hawary and his colleagues isolated two novel metabolites, named Saccharomonosporine A (
9) and convolutamydine F (
10), and three new compounds (
11,
12,
13) from the co-cultured extract of
Saccharomonospora sp. UR22 and
Dietzia sp. UR66 are both derived from the Red Sea sponge
Callyspongia siphonella [17]. The co-cultivation of marine wharf roach gut bacterial strains
Streptomyces sp. GET02.ST and
Achromobacter sp. GET02.AC induced the amount of the new natural product, ligiamycins A (
14), 24 times more than that only from GET02.ST in mono-culture. It also led to the discovery of a new natural product, ligiamycins B (
15) (
Figure 1)
[18]. Although endophytes are considered a promising resource for the discovery of superior co-culture strains, Knowles and his colleagues thought that the endophyte community depends not only on the host but also on the geographical location
[19].
Figure 1. The structures of compounds 1–15.
In order to survive in special environments such as hydrothermal and cold springs containing extreme pH, temperature, salinity, and pressure, microbial communities have developed unique defenses against their environment, leading to the biosynthesis of new molecules
[20]. In anaerobic environments, for example, different metabolic processes are either stimulated or dependent on other partners
[4][21]. The complexity of interactions among microbial communities growing in special environments makes them suitable co-culture partners. Examining microbial communities’ laboratory cultivability and interaction in special environments is a possible direction for enriching the natural product library.
2.2. Pairing with Pathogens
Globally, plant pathogens threaten a substantial portion of food production, with a 20–30% loss of crops estimated, primarily in food-deficit areas
[22]. Danquah et al. obtained 21 fungi via screened from 121 fungi which isolated from the samples of water, sediment, seabed foam, and plant materials (seeds, branches, leaves, sawdust) collected at the Windebyer Noor. They used these genetically diverse fungal strains to co-culture with two phytopathogenic bacteria (
Pseudomonas syringae and
Ralstonia solanacearum) and two phytopathogenic fungi (
Magnaporthe oryzae and
Botrytis cinerea), respectively, in two pre-selected solid media Sabouraud medium (SA) and Potato Dextrose Agar medium (PDA). The extracts of nine co-cultures showed biological inhibition ≥70% at a concentration of 200 μg·mL
−1, targeting at least three plant pathogenic test strains
[23]. To highlight these results, Danquah et al. chose to conduct a more in-depth study on the co-culture extracts of marine
Cosmospora sp. and phytopathogen
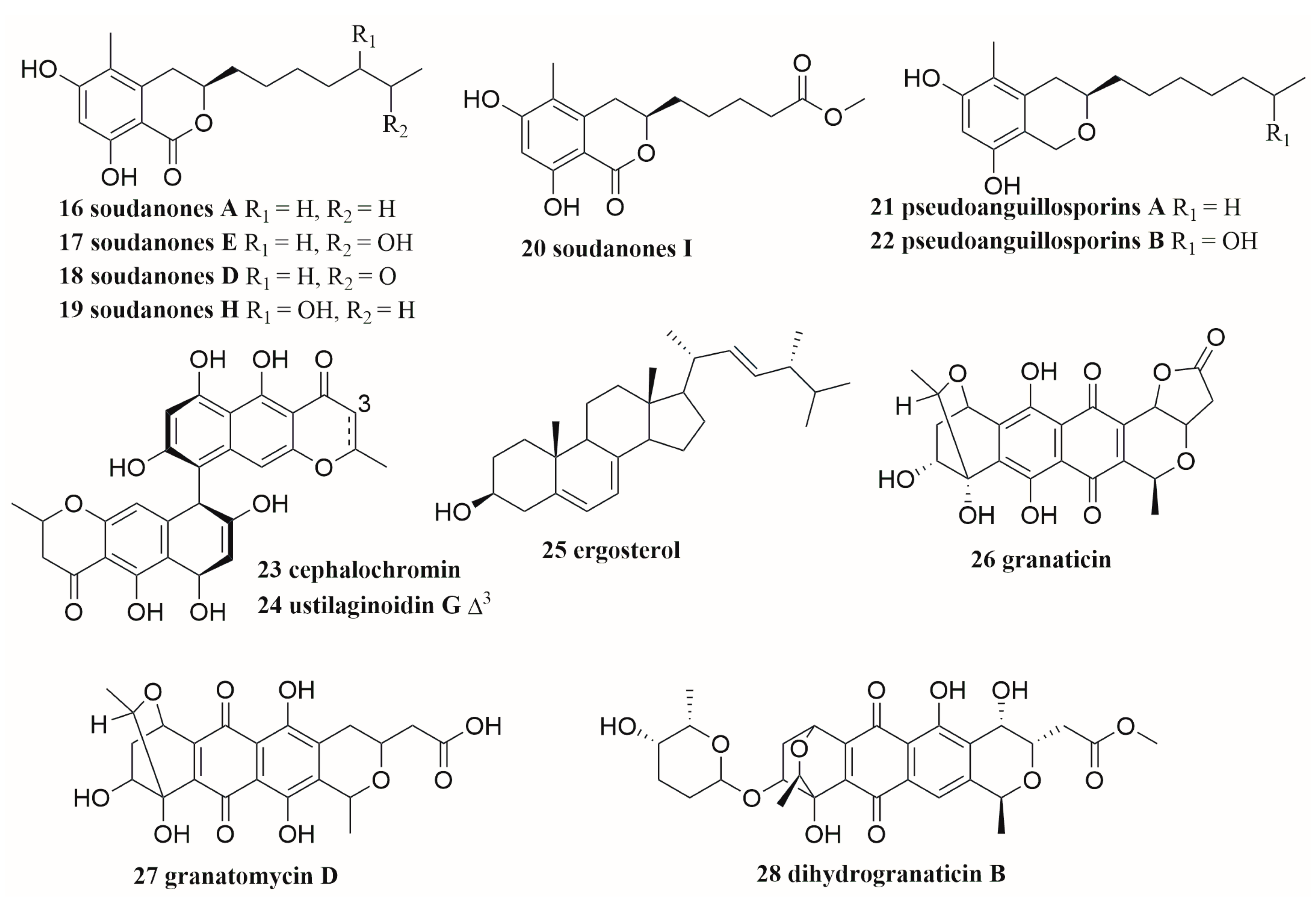
M. oryzae. Ten compounds, including two new isochroman-1-one derivatives, soudanones H-I (
19,
20) and their three known homologues (
16,
17,
18), and other five known compounds, namely pseudoanguillosporins A and B (
21,
22), cephalochromin (
23), ustilaginoidin G (
24), and ergosterol (
25), were identified from the extracts of large-scale co-culture of
C. sp. and
M. oryzae (
Figure 2)
[24]. In addition, they experimented with a new alternative co-culture method, which classified eight marine sediment-derived fungi (
Helotiales sp.,
Plenodomus influorescens,
Penicillium bialowiezense,
Sarocladium strictum,
Pyrenochaeta sp.,
Pyrenochaeta nobilis, and two
Lentithecium strains) as “strong” or “weak” categories based on their extraction-resistant abilities to six phytopathogens (
Erwinia amylovora,
P. syringae,
R. solanacearum,
Xanthomonas campestris,
M. oryzae, and
Phytophthora infestans). Then, the co-culture of the three modes was implemented, including pairings of weak–weak, strong–strong, and weak–strong. Finally, the co-culture extracts of
Plenodomus influorescens (strong), and
Pyrenochaeta nobilis (weak) showed significant differences in biological activity and metabolic profiles
[25].
Figure 2. The structures of compounds 16–28.
The presence of resistant pathogens can create a survival pressure on antibiotic-producing strains, forcing antibiotic-producing microorganisms to produce new antibiotics or up-regulate the original antibiotic, thus gaining a competitive advantage
[26]. Sung et al. co-cultured the tunicate-associated bacterium
Streptomyces sp. PTY087I2 with four human bacterial pathogens (
Bacillus subtilis, methicillin-sensitive
Staphylococcus aureus, methicillin-resistant
Staphylococcus aureus (MRSA), and
Pseudomonas aeruginosa). The molecular network of the co-culture extracts of
S. sp. with these human pathogens showed that the increased production of three antibiotics including granaticin (
26), granatomycin D (
27), and dihydrogranaticin B (
28) (
Figure 2), and several known analogues were discovered. The biological activity of co-cultured strain extracts against Gram-positive human pathogens used in these experiments was significantly increased
[27]. Three types of strains, namely heat-killed bacterial pathogens, living bacterial pathogens (
S. aureus, B. subtilis, E. coli, and
P. aeruginosa), and environmental strains (
Streptomyces and
Arthrobacter), were used to co-culture with
Streptomyces sp. AC117. The result showed that adding living pathogenic bacteria or phylogenetically closed bacteria to AC117 did not significantly improve the antibacterial activity, while co-cultured with heat-killed bacteria (especially for
S. aureus), the antibacterial activity of the extracts from AC117 was enhanced, as well as a five-fold increase for the yield of anthraquinones
[28]. Ozkaya et al. co-cultured five marine sponge-derived fungi, namely
Cladosporium sp. (SF17),
Penicillium Commune (SF42),
Aspergillus carneus (SF114B),
Penicillium canescens (SF146A), and
Aspergillus iizukae (SF153A), with the mixture of four fish pathogenic bacteria
Vibrio anguillarum O1,
Vagococcus salmoninarum,
Yersinia ruckeri, and
Lactococcus garvieae, respectively. The co-culture induced the increase in antibacterial activity of the two strains:
A. iizukae (SF153A) and
A. carneus (SF114B). The mixed fermentation extract of
A. iizukae showed strong inhibitory activity against
L. garvieae, with a minimal inhibitory concentration (MIC) value of 1.6 μg·mL
−1 [29].
2.3. Pairing with Sub-Inhibitory Concentration Antibiotics
Some work suggested that secondary metabolites with antibiotic activity might act as microbial defense molecules in natural microbial communities, and that they might also act as cues or signals to trigger gene expression and phenotypic changes at sub-inhibitory concentrations. In applying high-throughput screening on small molecular libraries to find potential inducers for activating silent gene clusters, Seyedsayamdost et al. proved the obvious indigenous activation of two silent gene (
Burkholderia thailandensis) clusters: the malleilactone cluster and the Burkholdac cluster. They revealed that most initiators themselves were antibiotics. Antibiotics would kill
B. thailandensis at high concentrations while acting as an inducer of secondary metabolism at low concentrations
[30]. Buijs et al. observed an expression of orphan biosynthetic gene clusters in
B. thailandensis when treated with sub-MIC andrimid which was produced by a marine bacterium
Vibrio corallilyticus. They tested the effect of sub-inhibitory concentrations of andrimid on another marine Vibrionaceae (
Photobacterium galatheae). The results showed that andrimid could stimulate the reporter strain
P. galatheae in a sub-MIC range, and that by UV–visible spectroscopy, andrimid at low concentrations could stimulate the increase in holomycin, a secondary metabolite of
P. galatheae. Furthermore, in the extract of andrimid-treated
P. galatheae, they observed a 10-fold up-regulation of an orphan biosynthetic gene cluster, which subsequently resulted in a 1.6–2.2 fold up-regulation of the biosynthesis of holomycin
[31].
2.4. Pairing with Phototrophs
The basic concept of ecology emphasizes nutrient cycling between phototrophic and heterotrophic organisms. Heterotrophic organisms usually promote the growth of phototrophic organisms in co-culture. This relationship is helpful to establish an effective microalgae–bacteria co-culture model
[32][33]. Chhun and his collaborators found that when phototrophs and marine actinobacteria were cultured together as partners for the first time, the photosynthate (released by photosynthetic primary producers) could activate biosynthetic gene clusters of marine actinobacterium
Salinispora tropica, including the orphan biosynthetic gene clusters
pks3 and
nrps1 [34].
2.5. Pairing with Mycolic Acid-Containing Bacteria
Bacteria containing mycolic acid were sometimes selected as paired strains because they could induce pigment production
[35]. When the
pks13 gene related to the biosynthesis of mycolic acid in
Tsukamurella pulmonis TP-B0596 was destroyed, the pigments induced by co-culture of
T. pulmonis TP-B0596 with
Streptomyces lividans TK23 were not observed
[36]. The co-culture analysis of marine-derived
Aspergillus niger and a mycolic acid-containing bacteria
Mycobacterium smegmatis suggested that a pigment produced by
A. niger was induced, and the cytotoxicity of crude extract on human prostate cancer DU145 cells was significantly increased compared with the mono-culture
[37].
2.6. Pairing with Microorganisms Containing Halogen Peroxidase-Related Gene Cluster
The high proportion of natural products containing bromine and a small number of chlorine in marine-derived natural products is a prominent characteristic of marine natural products
[38]. Halides in seawater are relatively abundant, and marine organisms can usually incorporate these elements into their metabolites through the catalysis of enzymes. However, the statistical results of secondary metabolites of marine-derived microorganisms were interesting. Compared with marine invertebrate natural products (10.1% containing bromine) and marine plant natural products (36.8% containing bromine), only 1.8% of marine microbial natural products contained bromine, and this value is only slightly higher than that (0.7%) which exists in terrestrial microorganisms. Voser et al. proposed that it might be attributed to the presence of a co-culture-like ecosystem around marine-derived microorganisms and microorganisms containing halogen peroxidase-related gene clusters, which promote the halogenation of metabolites
[38].
3. How to Select the Co-Culture Conditions?
3.1. Culture Parameters
3.1.1. Scale of Fermentation
Large-scale co-culture can be carried out on solid agar plates. However, this cultural method has some limitations when a large number of metabolites is needed for structural identification and bioassay. Liquid substrate can also be selected as the medium for the co-culture of strains, commonly known as mixed fermentation
[39]. The microscale co-culture method can promote the diversity of strain combinations, facilitate the evaluation of the repeatability, determine the scalability of the culture method, and provide a reference for large-scale fermentation. A total of 65 Micromonosporaceae and bacteria containing mycolic acid were co-cultured or cultured alone by microscale fermentation (500 μL). As a result, 12 Micromonosporaceae across three genera could produce unique metabolites during the co-culture
[40].
3.1.2. Optimization of Culture Parameters
Changes in the yield and biological activity of secondary metabolites were observed in the co-culture of autoclaved strains at different growing stages. Considering that both
Aspergillus. terreus C23-3 and
Aspergillus. unguis DLEP2008001 can produce effective antibiotics and other bioactive compounds, Wang et al. co-cultured the two strains continuously or simultaneously in a live or inactivation form, respectively, to detect the difference of metabolites. For example, later inoculated
A. terreus when co-cultured with autoclaved
A. unguis resulted in a diversity of metabolites, whereas pre-autoclaving
A. terreus effectively inhibited the growth and metabolism in
A. unguis. Moreover, the metabolites of
A. terreus still showed high stability after autoclaving. Their further study demonstrated that co-culture metabolites were usually dominant when the strains were pre-inoculated, and the yields could even be higher than those of mono-culture. Pre-inoculated strains strongly inhibit the growth and metabolism of subsequent inoculated co-culture strains by up-regulating or producing specific secondary metabolites
[41]. Jomori et al. compared the induction effects of autoclaved and living
M.
smegmatis on marine-derived
A. niger, and the results showed that the extracts of autoclaved
M. smegmatis co-cultured with
A. niger were not significantly different from those of the mono-culture
[37]. In 2020, Mokkala et al. co-cultured live or autoclaved
B. subtilis with five marine-derived fungi (
Eurotium chevalieri,
Emericella foveolate,
Myrothecium verrucaria,
Talaromyces tratensis, and
Talaromyces stipitatus), and compared the inhibitory effects of extracts on plant pathogens under different culture conditions. The results showed that co-culture with autoclaved
B. subtilis significantly reduced the antifungal activity of tested marine-derived fungi
[42]. During the co-culture, the growth rate and maximum cell density of the two strains
P. galatheae and
V. coralliilyticus were very sensitive to the inoculation rate, and different inoculation rates induced different degrees in stimulating the metabolites of
P. galatheae and
V. coralliilyticus [43]. Boruta et al. characterized the secondary metabolites and studied the biological process kinetics of
Aspergillus terreus ATCC and
Streptomyces noursei ATCC 11455. They compared the biosynthesis ability of the two strains in co-culture and in mono-cultures. In addition, they established another extensive data set, about the consumption of dissolved oxygen and the uptake rates of glucose and lactose. Generally, it is not surprising to obtain a co-culture model that showed almost no differences in product formation, substrate consumption, and morphological characteristics from the corresponding mono-culture. One of the key issues to be considered when planning co-culture experiments is the dominance of the more “aggressive” strains over their partners. The more “aggressive” strain’s dominance on its partner makes it win in the co-culture. It is essential to design a specific inoculation scheme, namely “Draw”, to prevent the overgrowth of the producing strain. In co-culture, the unexpected “Draw” between
A. terreus and
S. noursei was surprising. Compared with other co-culture groups, “Draw” was beneficial for the inoculation plan in stimulating the production of secondary metabolites. The inoculation scheme and medium composition were considered as essential aspects to affect the synergistic development and biosynthetic activity of the two strains. In addition, Boruta et al. designed a methodological approach to evaluate the growth status of co-cultured members by using substrates selectively metabolized in the individual species. In the microbial co-culture system of
S. noursei and
A. terreus, since
S. noursei was proven unable to use lactose, the change level in lactose could reflect the growth of
A. terreus [44]. In the early stages of co-culture development, lack of repeatability is a factor hindering its development. However, various studies have shown that co-culture can provide repeatable metabolite patterns when the relevant fermentation parameters are firstly optimized and carefully maintained. Wakefield and his collaborators conducted small-scale cultures of bacteria and fungi individually, as well as their co-cultures under different conditions. The fermentation parameters were optimized by liquid chromatography–high resolution electrospray ionization mass spectroscopy (LC-HRESIMS), liquid chromatography-ultraviolet (LC-UV), and microanalysis to maintain the reproducibility of metabolite production. Once the optimized fermentation parameters were determined, large-scale co-culture was conducted to isolate the natural products. The observed small-scale experiments were highly comparable to large-scale fermentations
[45]. Rateb et al. isolated 10 secondary metabolites from the co-culture extract of
Aspergillus fumigatus MBC-F1-10 and
Streptomyces bullii. In order to understand the possible mechanism of
Streptomyces inducing the biosynthesis of these fungal metabolites, the effects of medium composition, bacterial biomass extracted by MeOH, bacterial culture broth inactivated by autoclaving, and metabolism of the fungus MBC-F1-10 were studied
[46]. Five algicidal tryptamine derivatives were isolated from the co-culture broth of marine
Streptomyces and
Bacillus mycoides. In order to solve the low production rates and low reproducibility of metabolites in co-culture, factors such as nutrient composition, culture mode, and pH value of the medium were optimized. The methods of reducing the growth rate gap and slowing down the growth speed were used to prolong the co-culture time of two microorganisms, resulting in an increase in the yield of tryptamine derivatives
[47]. Different co-culture methods have been tried to elevate the induction rate of diketopiperazines, including the inoculation times, amounts of two microbes, and the water content of rice medium in the co-culture of the fungus
Penicillium sp. DT-F29 and the bacteria
Bacillus sp. B31
[48].
3.2. Culture Apparatus
A porous filter was used to physically separate
S. tropica and
Synechococcus strains during the co-culture, preventing direct intercellular interactions while allowing the diffusion of small molecules
[34]. In the co-culture of
Penicillium sp. LXY140-R and
Penicillium sp. LXY140-3, Li et al. designed a dialysis bag separation device, allowing only small molecules to penetrate. Co-culture studies showed that the LXY140-3 strain accumulated trichothecene sesquiterpenes by setting up a co-culture device
[49]. For the co-cultivation of the strain,
Streptomyces sp. MA37 and the Gram-negative bacterium
Pseudomonas sp., Maglangit and others developed a device consisting of two glass chambers connected by a holding steel clamp and a silicone O-ring. The two culture containers were separated with a polyvinylidene fluoride (PVDF) membrane filter, which permitted the two strains to grow under the same culture conditions but without direct contact. High-performance liquid chromatography (HPLC) traces showed that several unidentified metabolites were up-regulated or expressed in the extracts of co-cultured
P. sp. and
S. sp., compared with mono-culture
[39].
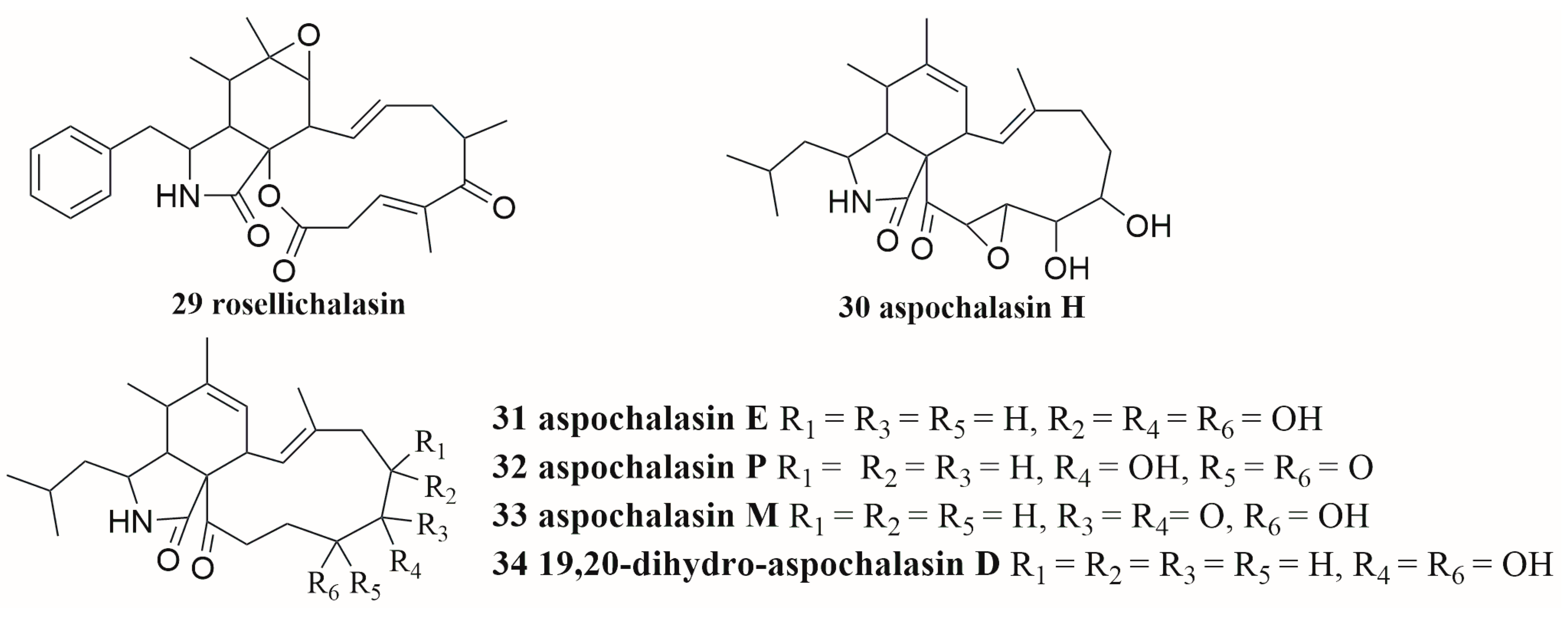
In some cases, the induction of secondary metabolites requires physical contact between paired strains. The co-cultures of
Aspergillus flavipes and
Streptomyces sp. resulted in the up-regulation of cytochalasans (
29–
34) (
Figure 3), a series of secondary metabolites of
A. flavipes. In a membrane-separated culture experiment, scanning electron microscopy morphological analysis showed that the effective induction of secondary metabolites required physical contact of microorganisms
[50]. In an in-depth study of the co-culture extract of marine-derived
Cosmospora sp. and phytopathogen
M. oryzae, Ernest et al. observed the induced isochromanones and their accumulation in the antagonistic zone in the overlaid co-culture on solid agar, suggesting that direct intercellular contact between
C. sp and
M. oryzae may be critical for isochromanones’ production on PDA media
[24]. The co-cultivation of
A. niger and
M. smegmatis led to the production of a pigment by
A. niger. The extract of co-culture exhibited increased cytotoxic activity against human prostate cancer DU145 cells. Pigment would be produced by
A. niger and not be observed in an
A. niger and
M. smegmatis combined culture mode treated in two separated compartments partitioned with a dialysis membrane. The cytotoxicity of the extract was not significantly different from that in axenic culture of
A. niger. Similarly, the extracts of autoclaved
M. smegmatis co-cultured with
A. niger were not significantly different from those of the mono-culture. Therefore, it was necessary to co-culture living
M. smegmatis and
A. niger in cell–cell interaction
[37].
Figure 3. The structures of compounds 29–34.