1. Introduction
A mass spectrometer consists of at least three components: a desorption/ionization source, mass analyzer, and ion detector. The mass analyzer and ion detector in mass spectral imaging (MSI) are under a vacuum; some desorption/ionization sources are a under vacuum, while others operate at atmospheric or intermediate pressures
[1]. Ion generation is accomplished when material from the sample surface is volatilized and ionized into the gas phase. This process can be performed in multiple ways. Once the ions are created, they then move through the mass analyzer to the mass detector, where the ion signal is converted into an electrical signal. The mass spectrum is generated by the interpretation of the electrical signal generated
[2].
Mass spectrometry has advanced significantly within the past decade, and it is one of the most widely used analytical platforms. Among MS techniques, MSI or imaging mass spectrometry has advanced the field further and led to wide applications in geological, biological, and medicinal research
[3][4]. Matrix-assisted laser desorption ionization (MALDI) mass spectrometry and secondary ion mass spectrometry (SIMS) are two of the leading MSI techniques used in investigations in biology and microbiology. Other techniques, such as desorption electrospray ionization (DESI), laser ablation electrospray ionization (LAESI), laser desorption postionization (LDPI)
[5], and secondary neutral ionization (SNMS), are also used in biology and microbiology research
[3][6][7][8][9][10].
In MSI, an image is created by collecting spectra from different parts of the sample surface in succession. This can be achieved by moving the target specimen so that the desorption/ionization source may examine or investigate different sections of the sample. Moving of the sample is typically accomplished by mounting the sample on a motorized stage, movement of the sample using a piezo stage, manually, or by translating the desorption/ionization probe (i.e., focused laser or ion beam) while the sample remains in place. Once ions are generated, they are extracted into the mass analyzer and detected using a mass analyzer and ion detector, which can vary with the specific configuration of the instrument. Unlike MS used in the analysis of bulk samples, each mass spectrum collected for individual spots analyzed corresponds to pixels that are used in mass spectral images. A range of pixels can be chosen in MSI data acquisition. The detection area, or image area, is also defined by the user and is set before experiments. Rastering over a large area and having smaller spaces between pixel generation will lead to the development of a spatially resolved image that contains sample specific information
[6]. Private and public source software allows users to interact with these data by assigning relative distributions to a map of mass spectra for visualization, containing different intensities of mass-to-charge (
m/
z) ratios
[6].
MSI has become an important tool for biologists and microbiologists alike over the past two decades due to the molecular information collected and contained within the spectral images
[11][12]. MSI is powerful because it offers enhanced visualization and spatial resolutions, offering specificity, sensitivity, and temporal information
[6][7][13].
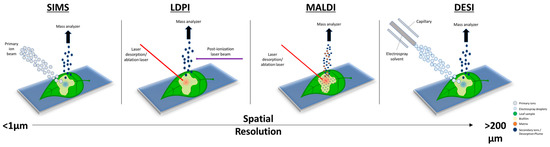
Figure 1 shows the main MSI techniques and their corresponding spatial resolutions. Plant–microbe interactions have evaded intact molecular analyses for many years due to the nature and scale in which these interactions occur. MSI, on the other hand, can offer a solution to most, if not all, of the problems encountered with more traditional approaches. A growing concern, as population growth continues to rise, is biomass production and crop sustainability, which is a subject that has a close dependence on plant–microbe interactions. It is estimated that >100 bacterial cells exist per gram of soil
[14]. Bacteria form microbiomes for the surrounding plant species, typically colonizing the rhizosphere, rhizoplane, phyllosphere, spermosphere, and endosphere
[14][15]. The plant–microbe interactions can either be mutualistic or parasitic depending on the plant–soil feedback
[16]. Mapping the biological metabolic pathways and understanding the interactions that microbes have with plants at the molecular level is possible with the use of MSI. The information gained, either quantitative or qualitative, can be far more valuable and relevant compared to standard mass spectrometry measurements of a homogeneous bulk sample
[17].
Figure 1. Overview of the different desorption/ionization methods used for mass spectrometry imaging (MSI) to map plant–microbe interactions. The scale bar shows the spatial resolution of each technique, from sub-micrometer (μm) to greater than 200 μm. A higher spatial resolution corresponds to higher-quality mass spectral images.
MSI has been used to study plant–bacteria interactions and gained more interest in recent years. Specifically, there have been approximately 35 articles covering this topic of plant–microbe interactions since 2004 and six reviews since 2014, according to Web of Science survey results. Confocal laser scanning microscopy (CLSM) is the primary method used for imaging plant–microbe interactions due to its ease of access and reasonable spatial information
[18]. Boughton et al.’s review of MSI for plant biology offered a comprehensive overview of MSI capabilities and advances made with the different imaging techniques up to 2015
[3]. Musat et al. tracked microbial interactions with different hosts, plant or animal, via NanoSIMS in a review in 2016
[19]. In 2017, Ho et al. covered the different applications of MSI and detailed microbe–microbe interactions as well as plant–microbe interactions
[20]. While MSI has not been a mainstream method for the study of plant–microbe interactions, MSI can provide insights into metabolic pathways, metabolite identification, plant-growth-promoting bacteria, and biotic stress.
2. Secondary Ion Mass Spectrometry
Secondary ion mass spectrometry (SIMS) is a method that uses a primary ion beam to desorb analytes from the sample surface, in turn creating a plume of secondary ions and neutrals. While there are many configurations of SIMS instruments, the fundamental basis remains the same: the measurement of the mass and intensity of secondary ions produced while under a vacuum after sputtering the sample surface with an ion beam or neutral beam
[21]. Early experiments with ion desorption can be traced back to R. F. K. Herzog and F. P. Viehbock, where they experimented with new ion sources and the sputtering of a sample. Then, in 1957, D.G. Bills demonstrated the desorption of ions from metal surfaces after the bombardment of the metal surface with nitrogen ions or low-energy electrons, stemming from earlier works by Herzog
[22]. In 1963, the sputtering ion source using concentrated argon ions to bombard the sample surface for secondary ion desorption was introduced by Liebl and Herzog
[23]. However, it was not until 1972, when Huber, Selhofer, and Benninghoven demonstrated that the SIMS technique under an ultra-high vacuum (UHV) was able to measure both positive and negative ions, and to acquire profiles of multilayer films, that the basis for the modern SIMS instruments was set
[24]. SIMS imaging was not far behind after sputtering sources were announced, and early SIMS imaging could be attributed to the ion microscope of a special design, which was used to image a surface region of less than 0.5 mm
2 [25].
SIMS has been at the forefront of the development of MSI techniques for decades. Levi-Setti et al. published an article on the progress of high-resolution scanning ion microscopy and SIMS imaging microanalysis in 1985. Kingham et al. published their work on three-dimensional SIMS imaging and the depth profiling of biological material, such as bone tissue, in 1987, showing that the ToF mass analyzer was best for imaging when compared with a quadrupole or magnetic sector due to its greater sensitivity. However, the erosion time was several orders of magnitude less, thus limiting this method to thin films and small area depth profiles
[26]. One of the earliest publications on plant–microbe interactions was from Lorquin et al., where they used HPLC and GC-MS to determine nodule factors from soil bacteria interacting with host plants
[27]. Since then, ToF-SIMS and another method known as nanoSIMS (see below) have been used for the imaging of plants, bacteria, and plant–microbe interactions, to analyze carbon and nitrogen assimilation by soil microbes
[28], map nutrient uptake in situ in the rhizosphere
[29], and image the in situ flow of photo-assimilated carbon through arbuscular mycorrhiza into root and hyphae-associated soil microbial communities
[30].
More recently, Zhang et al. showed that ToF-SIMS imaging could be used to study the effects of plant-growth-promoting rhizobacteria on
Brachypodium awn. Plant-growth-promoting rhizobacteria reside within the rhizosphere
[31].
Brachypodium distachyon, a C3 representative plant model, has been used to show plant–microbe interactions, and it is considered an ideal species for this type of experiment
[31][32]. Zhang et al. studied the interaction of
Arthrobacter chlorophenolicus and
Pseudomonas fluorescens, planktonic cells and biofilms on the
Brachypodium seed, which are referred to as awns. They were able to characterize plant metabolites, such as flavonoids, phenolic acids, fatty acids, and indole-3-acetic acid, on the awn surface. They also used principal component analysis (PCA) for the evaluation of the characterized metabolites and their interactions with the awn surface. The experiment determined that certain fragments of flavonoids, such as kaempferol (
m/
z− 285, C
15H
9O
6−) and quercetin (
m/
z− 301, C
15H
9O
7−), were present in only the
Pseudomonas-treated samples and suggested that flavonoids respond more against pathogenic Gram-negative species
[31]. They also showed that fatty acids’ observed intensity increased when plant-growth-promoting bacteria were introduced to the seedling or awn. The PCA results determined that both Gram-negative and Gram-positive bacteria affected the awns.
3. Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry
MALDI mass spectrometry is a laser desorption ionization technique that involves treating the sample with a matrix substance, either solid or liquid, to enhance the ionization of the sample. The matrix is typically compounds such as dihydroxybenzoic acid (DHB)
[33], sinapinic acid (SA)
[34], or α-cyano-4-hydroxycinnamic acid (CHCA)
[35]. MALDI has been used in several publications involving microbial interactions and it can provide information on the molecular species of the microbial communities
[36]. However, MALDI analysis is highly dependent on the selection of the correct matrix for the sample, how well the laser interacts with the matrix, and the details of the sample preparation prior to analysis. It is critical when selecting the matrix that one understands the absorption range so that it is compatible with the laser. Incompatible matrix treatment will provide little to no useful ion signal from the sample. The absence of ion signal can result from insufficient desorption and/or ionization of the analyte, and one often does not know the reason for the failure. Additionally, it is crucial when a matrix is applied that there is co-crystallization with the sample, so that the laser pulses will desorb and ionize not only the matrix but the sample molecules as well
[34]. Co-crystallization provides an equal distribution of sample and matrix and allows the laser to easily desorb the material from the sample surface, thus increasing the ionization and ion yield. CHCA is typically used for small molecules and peptide samples, whereas DHB is used for lipids. The latter can be used for small molecules and SA is primarily used for protein analysis
[17].
MALDI is most commonly coupled to a ToF mass analyzer, which works by extracting the ions generated under a vacuum in the sample chamber and introducing them into the flight tube. Here, ions can be steered and directed with ion optics such as Einzel lenses while varying the applied voltages. As the ions travel through the flight tube, they will begin to separate based on their mass and charge ratios (
m/
z), leading smaller, lighter
m/
z ions to reach the detector first and the larger, heavier ions to follow. Orbitrap mass analyzers are also used in MALDI
[37][38][39][40][41]. Orbitraps operate by trapping an ion radially around a centralized spindle electrode. As the trapped ions rotate around the spindle electrode, independent of the energy and spatial spread, the harmonic oscillations displayed as electromagnetic radiation are measured and converted into
m/
z values by Fourier transforms
[42]. Orbitrap mass analyzers have high resolving power—for example ~160,000 at
m/
z 750—and mass accuracy, which enhances the chemical information obtained from the mass spectra
[38].
MALDI MSI has been used for the quantification and spatial localization of organic acids in root exudates. Low-molecular-weight organic acids from root exudates were examined for their roles in plant nutrition and pH modification in the rhizosphere, and as bio-stimulants and chemoattractants, by Gomez Zepeda et al.
[43]. Organic acid exudation from the plant roots to the rhizosphere was previously determined as a mechanism by which plants cope with cation toxicity. Creating methods for the rapid and precise quantification and localization of the organic acids in the plant roots has improved the understanding of their role in response to phosphate deficiency and toxic cations
[43].
MALDI was used to determine the spatial distribution and localization of malate and citrate within the plant roots of
Arabidopsis wild-type seedlings, aluminum activated malate transporter 1 (OX.ALMT1), and positive transcriptional regulator sensitive to proton rhizotoxicity 1 (OX.STOP1), which are the transgenic lines that the
Arabidopsis genome expresses. It was determined that the wild-type malate signal had no distribution differences in phosphate abundance or restriction. OX.ALMT1 and OX.STOP1 tended to have a wider spread in the rhizosphere and higher intensity with both phosphate abundance and restriction compared to the wild type, corroborating the findings that the overexpression of ALMT1 and STOP1 enhances malate exudation from the roots
[43].
4. Laser Desorption/Ionization Mass Spectrometry and Related Methods
LDI mass spectrometry has been around since the early 1960s, shortly after the invention of the laser in the 1950s. Levine, J. F. Ready, E. Bernal, W. I. Linlor, R. E. Honig, and J. R. Woolston were some of the researchers who first explored desorption plume dynamics and used this technique to develop high-sensitivity mass spectrometers
[44][45][46][47][48][49]. As time passed and the adaptations and improvements to LDI developed, more researchers began to use it and, incidentally, it became the foundation of MALDI after early experiments from Hillenkamp et al.
[50]. Since then, laser desorption has developed into an enormous field, with areas of interest in microbial and biofilm analysis
[36][51][52][53][54], elemental and molecular analysis for biomaterials
[55][56][57][58][59], cellular analysis
[60][61], drug analysis
[9][62], geological analysis
[4][63], and elemental analysis and instrumentation design
[64][65]. LDI is achieved by exceeding a substance’s ablation/ionization threshold, albeit by a mechanism still subject to debate: thermal vaporization, nonthermal melting, electron–lattice heating, shockwave propagation, plasma expansion, and proton or cation transfer have all been discussed as LDI mechanisms
[66].
Laser desorption has prospered since its inception and many different methods and adaptations have become available. A key development is laser desorption postionization (LDPI). It employs two nanosecond (ns) or femtosecond (fs) pulsed laser systems for separate desorption and photoionization steps. For example, fs laser desorption postionization mass spectrometry (fs-LDPI-MS) utilizes laser pulses <100 fs for the “cold” ablation or thermal vaporization of a sample, meaning that as the energy excites the atom or molecule, the electrons will release before the energy relaxation event, allowing this to be a universal ablation method causing minimal sample damage. The desorption laser releases primary ions and neutrals into a plume and a second vacuum ultraviolet (VUV) ns laser is used to ionize the neutrals within the plume, creating positions, while the primary ions are suppressed via a biased voltage grid
[4][5]. The postions are then introduced to the mass analyzer for analysis. LDPI has been used for microbial and biofilm analyses
[51][52] as well as geological analyses
[4].
Hieta et al. 2021 used laser ablation atmospheric pressure photoionization mass spectrometry (LAAPPI-MS) imaging with a 70 µm lateral resolution to analyze
Arabidopsis thaliana’s leaf trichome and vein structures for metabolite identification and mapping
[60]. LAAPPI-MS utilizes an ns pulsed mid-IR laser, the same as used in LAESI, to ablate the leaf surface to generate a plume that is intercepted by the nebulized solvent spray and photoionized by a VUV lamp, both directed towards the mass analyzer inlet. A portable version of LAAPPI-MS was demonstrated using a handheld laser and an ion trap mass spectrometer
[67].
Hieta et al. expanded the LAAPPI-MS strategy to image single trichome cells and map the trichome base, leading to the mapping of different metabolite compounds, such as kaempferol, quercetin, isorhamnetin, rhamnose, and glucose
[60]. They acquired mass spectra for many lipids within the leaf lamina region and they differed from the trichome region. Most noticeably, they were able to show that LAAPPI-MS can perform the accurate depth profiling of plant material and accurately spatially resolve different structures at different depths within the leaf. Not only do the spectra results show that there is a difference between depth profile peak scans P1 and P2, but the images show that trichomes can be clearly seen at different depths at
m/
z 423.42, which further promotes this as a suitable imaging technique.
5. Electrospray Ionization, Desorption Electrospray Ionization, and Laser Ablation Electrospray Ionization Mass Spectrometry
Electrospray ionization (ESI) is the most widely used ionization method for non-volatile organic and biomolecules that are introduced by a liquid feed by direct injection or a liquid chromatograph
[68]. The liquid sample is introduced into a hypodermic needle held at a high voltage under atmospheric pressure, and the resulting field at the tip of the needle disperses the sample as a charged spray driven by Columbic forces
[69]. The charged spray passes into a mass spectrometer interface containing ion optical components and a series of stages of reduced pressure ending in the mass analyzer. A counter current bath gas flowing around the hypodermic needle is used to expedite the charged droplet’s evaporation, causing the droplet to become smaller, leading to an increased surface charge, as the droplets and ions then pass into a vacuum. Once the critical Rayleigh limit is reached, a Coulomb explosion event takes place, shredding the droplet and producing smaller droplets. They are sometimes referred to as daughter droplets, which then evaporate and create quasi-ions that can be analyzed
[52][69]. ESI is considered a soft ionization technique since it leads to less sample fragmentation than electron impact ionization (used on gas feeds from a gas chromatograph).
Desorption electrospray ionization (DESI) impinges a steam of highly charged solvent droplets from an ESI source onto a solid sample: the scattered species containing the analyte are then captured by the mass spectrometry interface
[70][71]. Samples analyzed by DESI are held at atmospheric pressure, whereas MALDI, SIMS, and LDI/LDPI are operated typically under a vacuum
[13]. DESI has become increasingly used for the analysis of the plant rhizosphere since it is not limited to sample preparation for vacuum analysis. However, DESI has a relatively poor lateral resolution when compared to the other ionization techniques
[57][68]. Laser ablation electrospray ionization (LAESI) was developed by Vertes and coworkers by combining a mid-infrared pulsed laser that excites water in a sample, leading to the ablation of neutrals in a plume
[72]. The ablation plume then crosses an electrospray flow, which postionizes the neutrals in a manner similar to traditional ESI. LAESI is conceptually like MALDI, except that water inherent in the sample acts as the matrix. LAESI has a higher lateral resolution than DESI, but a lower lateral resolution than MALDI. A variant is laser ablation atmospheric pressure photionization (LAAPPI), which uses a different method for the postionization of the neutrals in the ablation plume
[57][68]. Plant analysis is amenable to LAESI and LAAPPI, but the signal can vary with the water content (i.e., a lower signal is expected from regions of samples with higher cellulosic content).
Taylor et al. used high-resolution MSI to obtain 40 µm spatial resolution images of
Fittonia argyroneura leaves, and they were able to identify chemical species specific to the physical structure within the plant leaf
[10]. The mass detector used for this experiment was an Orbitrap, which allowed for a mass resolution of ~60,000 but only slow image acquisition times. Taylor et al. were able to desorb and analyze samples from distances of 6–80 mm. The
Fittonia argyroneura leaf was sectioned into 1.5 cm
2 pieces, which included the veins and mesophyll, for MSI detection. The analysis produced metabolite-rich spectra and identified catechol, furoic acid, phthalide, lysine, and glycinamide ribonucleotide.