
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Leszek Kalinowski | -- | 3471 | 2023-08-17 10:23:18 | | | |
| 2 | Lindsay Dong | Meta information modification | 3471 | 2023-08-17 11:16:56 | | |
Video Upload Options
Skin diseases such as psoriasis (Ps) and psoriatic arthritis (PsA) are immune-mediated inflammatory diseases. Overlap of autoinflammatory and autoimmune conditions hinders diagnoses and identifying personalized patient treatments due to different psoriasis subtypes and the lack of verified biomarkers. Proteomics and metabolomics have been intensively investigated in a broad range of skin diseases with the main purpose of identifying proteins and small molecules involved in the pathogenesis and development of the disease.
1. Introduction
2. Rodents as a Model for Psoriasis
2.1. Spontaneous Mouse Models
2.2. Induced Modulation of the Skin Environment
2.3. Psoriatic Human Skin Xenograft Model
2.4. Transgenic Models
2.5. T-Cell Transfer Rodents Model
2.6. Genome Editing-Models Based on CRISPR/Cas9 Technology
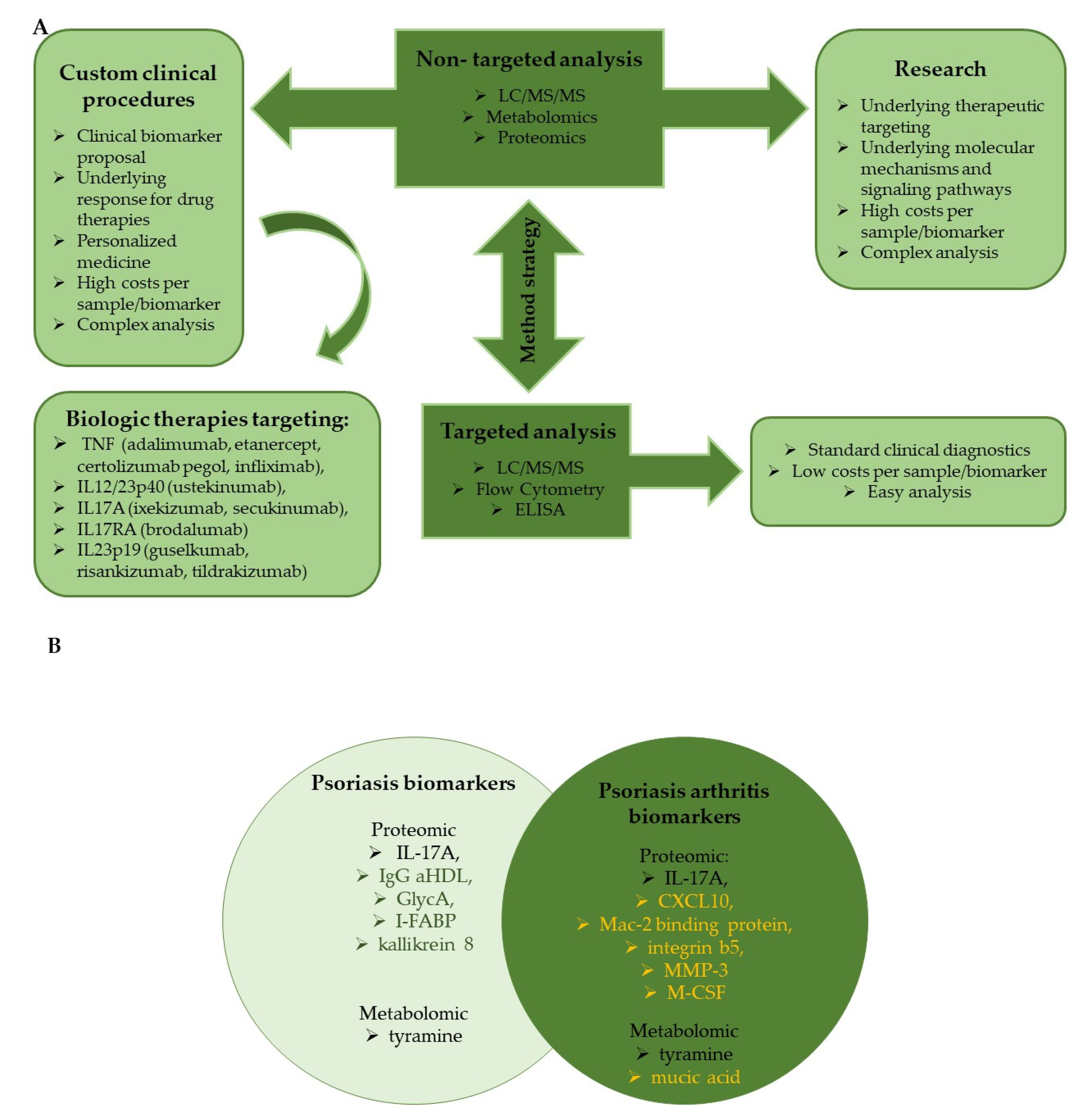
3. Proteomic and Metabolomic in Biomarker Discovery
3.1. Post-Translational Modifications (PTMs) of Proteins
3.2. Biological Material Selection for Proteomics/Metabolomics Studies
4. Clinical Trials in Proteomic and Metabolomic Biomarkers Discovery
Many of the selected clinical trials focused on biomarkers that could have the ability to predict responses to treatment with biologic disease modifying antirheumatic drugs (DMARDs), such as secukinumab, apremilast, and adalimumab. The primary purpose of the trials was to investigate inflammatory pathways and report changes in the levels of inflammatory markers at baseline and after treatment. To achieve the assumed goal, various analytical methods were used, such as RT-PCR and qPCR to assess the gene inflammatory biomarkers, flow cytometry to reveal the aberrant inflammatory profiles of cells, and histological examination to evaluate the reversing of lesion skin inflammation. The association between putative biomarkers and the progression and severity of psoriasis and psoriatic arthritis was investigated in several trials. Three independent clinical trials have identified matrix metalloproteinase (MMP-3) as a potential biomarker. This finding is consistent with the results presented by Ramessur et al., who identified MMP-3 as a proteomic candidate for predicting the development of psoriatic arthritis (PsA) from psoriasis lesions based on an analysis of 181 scientific articles [70]. Serum level of cytokines implicated in the Th17 pathway was measured in three clinical trials, indicating that IL 17A was a candidate for predicting psoriasis severity. In the study ‘Identification of New Prognostic Markers in Psoriatic Arthritis’, the concentration of IL 17A and other cytokines was correlated with markers of bone remodeling, identifying the molecular pathways involved in psoriatic arthropathy. The progression of psoriasis leads to the development of many comorbidities other than psoriatic arthritis, such as metabolic syndrome and cardiovascular diseases [78]. The study ‘Psoriasis Inflammation and Systemic Co Morbidities’ was intended to explore the pathophysiology of psoriasis and its comorbidities, but it also provided guidance on how long-term treatment of inflammation can reduce or prevent cardiovascular events.
5. Conclusions
References
- Yan, D.; Gudjonsson, J.E.; Le, S.; Maverakis, E.; Plazyo, O.; Ritchlin, C.; Scher, J.U.; Singh, R.; Ward, N.L.; Bell, S.; et al. New Frontiers in Psoriatic Disease Research, Part I: Genetics, Environmental Triggers, Immunology, Pathophysiology, and Precision Medicine. J. Investig. Dermatol. 2021, 141, 2112–2122.e3.
- Rendon, A.; Schäkel, K. Psoriasis Pathogenesis and Treatment. Int. J. Mol. Sci. 2019, 20, 1475.
- Zachariae, H. Prevalence of Joint Disease in Patients with Psoriasis: Implications for Therapy. Am. J. Clin. Dermatol. 2003, 4, 441–447.
- Bu, J.; Ding, R.; Zhou, L.; Chen, X.; Shen, E. Epidemiology of Psoriasis and Comorbid Diseases: A Narrative Review. Front. Immunol. 2022, 13, 880201.
- Griffiths, C.E.M.; Armstrong, A.W.; Gudjonsson, J.E.; Barker, J.N.W.N. Psoriasis. Lancet 2021, 397, 1301–1315.
- Sanchez, D.P.; Sonthalia, S. Koebner Phenomenon; StatPearls Publishing: Treasure Island, FL, USA, 2023.
- Gates, A.H.; Karasek, M. Hereditary Absence of Sebaceous Glands in the Mouse. Science 1965, 148, 1471–1473.
- Sundberg, J.P.; Dunstan, R.W.; Roop, D.R.; Beamer, W.G. Full-Thickness Skin Grafts from Flaky Skin Mice to Nude Mice: Maintenance of the Psoriasiform Phenotype. J. Investig. Dermatol. 1994, 102, 781–788.
- Sundberg, J.P.; Beamer, W.G.; Shultz, L.D.; Dunstan, R.W. Inherited Mouse Mutations as Models of Human Adnexal, Cornification, and Papulosquamous Dermatoses. J. Investig. Dermatol. 1990, 95, 62S–63S.
- Sundberga, J.P.; Francea, M.; Boggessa, D.; Sundberga, B.A.; Jenson’, A.B.; Beamera, W.G.; Shultza, L.D.; Words, K. Development and Progression of Psoriasiform Dermatitis and Systemic Lesions in the Flaky Skin (Fsn) Mouse Mutant. Pathobiology 1997, 65, 271–286.
- Brown, W.R.; Hardy, M.H. A Hypothesis on the Cause of Chronic Epidermal Hyperproliferation in Asebia Mice. Clin. Exp. Dermatol. 1988, 13, 74–77.
- Fallon, P.G.; Sasaki, T.; Sandilands, A.; Campbell, L.E.; Saunders, S.P.; Mangan, N.E.; Callanan, J.J.; Kawasaki, H.; Shiohama, A.; Kubo, A.; et al. A Homozygous Frameshift Mutation in the Mouse Flg Gene Facilitates Enhanced Percutaneous Allergen Priming. Nat. Genet. 2009, 41, 602–608.
- Wilkinson, D.I.; Karasek, M.A. Skin Lipids of a Normal and Mutant (Asebic) Mouse Strain. J. Investig. Dermatol. 1966, 47, 449–455.
- Vandeghinste, N.; Klattig, J.; Jagerschmidt, C.; Lavazais, S.; Marsais, F.; Haas, J.D.; Auberval, M.; Lauffer, F.; Moran, T.; Ongenaert, M.; et al. Neutralization of IL-17C Reduces Skin Inflammation in Mouse Models of Psoriasis and Atopic Dermatitis. J. Investig. Dermatol. 2018, 138, 1555–1563.
- Chan, T.C.; Hawkes, J.E.; Krueger, J.G. Interleukin 23 in the Skin: Role in Psoriasis Pathogenesis and Selective Interleukin 23 Blockade as Treatment. Ther. Adv. Chronic Dis. 2018, 9, 111–119.
- van der Fits, L.; Mourits, S.; Voerman, J.S.A.; Kant, M.; Boon, L.; Laman, J.D.; Cornelissen, F.; Mus, A.-M.; Florencia, E.; Prens, E.P.; et al. Imiquimod-Induced Psoriasis-Like Skin Inflammation in Mice Is Mediated via the IL-23/IL-17 Axis. J. Immunol. 2009, 182, 5836–5845.
- Nakahara, T.; Kido-Nakahara, M.; Ulzii, D.; Miake, S.; Fujishima, K.; Sakai, S.; Chiba, T.; Tsuji, G.; Furue, M. Topical Application of Endothelin Receptor a Antagonist Attenuates Imiquimod-Induced Psoriasiform Skin Inflammation. Sci. Rep. 2020, 10, 9510.
- Mohammed, S.S.; Kadhim, H.M.; Al-Sudani, I.M.; Musatafa, W.W. Anti-Inflammatory Effects of Topically Applied Azilsartan in a Mouse Model of Imiquimod-Induced Psoriasis. Int. J. Drug Deliv. Technol. 2022, 12, 1249–1255.
- Yang, Y.; Zhao, Y.; Lai, R.; Xian, L.; Lei, Q.; Xu, J.; Guo, M.; Xian, D.; Zhong, J. An Emerging Role of Proanthocyanidins on Psoriasis: Evidence from a Psoriasis-Like Mouse Model. Oxid. Med. Cell. Longev. 2022, 2022, 5800586.
- Schafer, P.H.; Chen, P.; Fang, L.; Wang, A.; Chopra, R. The Pharmacodynamic Impact of Apremilast, an Oral Phosphodiesterase 4 Inhibitor, on Circulating Levels of Inflammatory Biomarkers in Patients with Psoriatic Arthritis: Substudy Results from a Phase III, Randomized, Placebo-Controlled Trial (PALACE 1). J. Immunol. Res. 2015, 2015, 906349.
- Svensson, L.; Røpke, M.A.; Norsgaard, H. Psoriasis Drug Discovery: Methods for Evaluation of Potential Drug Candidates. Expert Opin. Drug Discov. 2012, 7, 49–61.
- Haftek, M.; Ortonne, J.P.; Staquet, M.J.; Viac, J.; Thivolet, J. Normal and Psoriatic Human Skin Grafts on “nude” Mice: Morphological and Immunochemical Studies. J. Investig. Dermatol. 1981, 76, 48–52.
- Raychaudhuri, S.; Raychaudhuri, S. Scid Mouse Model of Psoriasis: A Unique Tool for Drug Development of Autoreactive T-Cell and TH-17 Cell-Mediated Autoimmune Diseases. Indian J. Dermatol. 2010, 55, 157.
- Di Domizio, J.; Conrad, C.; Gilliet, M. Xenotransplantation Model of Psoriasis. In Inflammation: Methods in Molecular Biology; Clausen, B., Laman, J., Eds.; Humana Press: New York, NY, USA, 2017; Volume 1559, pp. 83–90.
- Tiirikainen, M.L.; Woetmann, A.; Norsgaard, H.; Santamaria-Babí, L.F.; Lovato, P. Ex Vivo Culture of Lesional Psoriasis Skin for Pharmacological Testing. J. Dermatol. Sci. 2020, 97, 109–116.
- Norsgaard, H.; Svensson, L.; Hagedorn, P.H.; Moller, K.; Olsen, G.M.; Labuda, T. Translating Clinical Activity and Gene Expression Signatures of Etanercept and Ciclosporin to the Psoriasis Xenograft SCID Mouse Model. Br. J. Dermatol. 2012, 166, 649–652.
- Gourlay, W.A.; Chambers, W.H.; Monaco, A.P.; Maki, T. Importance of natural killer cells in the rejection of hamster skin xenografts. Transplantation 1998, 65, 727–734.
- Ashkar, A.A.; Di Santo, J.P.; Croy, B.A. Interferon γ Contributes to Initiation of Uterine Vascular Modification, Decidual Integrity, and Uterine Natural Killer Cell Maturation during Normal Murine Pregnancy. J. Exp. Med. 2000, 192, 259–270.
- Chen, L.; Deshpande, M.; Grisotto, M.; Smaldini, P.; Garcia, R.; He, Z.; Gulko, P.S.; Lira, S.A.; Furtado, G.C. Skin Expression of IL-23 Drives the Development of Psoriasis and Psoriatic Arthritis in Mice. Sci. Rep. 2020, 10, 8259.
- Van Nuffel, E.; Staal, J.; Baudelet, G.; Haegman, M.; Driege, Y.; Hochepied, T.; Afonina, I.S.; Beyaert, R. MALT 1 Targeting Suppresses CARD 14-induced Psoriatic Dermatitis in Mice. EMBO Rep. 2020, 21, e49237.
- Schonthaler, H.B.; Huggenberger, R.; Wculek, S.K.; Detmar, M.; Wagner, E.F.; Karin, M. Systemic Anti-VEGF Treatment Strongly Reduces Skin Inflammation in a Mouse Model of Psoriasis. Proc. Natl. Acad. Sci. USA 2009, 106, 21264–21269.
- Retser, E.; Schied, T.; Skryabin, B.V.; Vogl, T.; Kanczler, J.M.; Hamann, N.; Niehoff, A.; Hermann, S.; Eisenblätter, M.; Wachsmuth, L.; et al. Doxycycline-Induced Expression of Transgenic Human Tumor Necrosis Factor α in Adult Mice Results in Psoriasis-like Arthritis. Arthritis Rheum. 2013, 65, 2290–2300.
- Voskas, D.; Jones, N.; Van Slyke, P.; Sturk, C.; Chang, W.; Haninec, A.; Olya Babichev, Y.; Tran, J.; Master, Z.; Chen, S.; et al. A Cyclosporine-Sensitive Psoriasis-Like Disease Produced in Tie2 Transgenic Mice. Am. J. Pathol. 2005, 166, 843–855.
- SchÖn, P.; Detmar, M.; Parker’, C.M. Murine Psoriasis-like Disorder Induced by Naive CD4+ T Cells. Nat. Med. 1997, 3, 183–188.
- Davenport, C.M.; Mcadams, H.A.; Kou, J.; Mascioli, K.; Eichman, C.; Healy, L.; Peterson, J.; Murphy, S.; Coppola, D.; Truneh, A. Inhibition of Pro-Inf Lammatory Cytokine Generation by CTLA4-Ig in the Skin and Colon of Mice Adoptively Transplanted with CD45RB Hi CD4 + T Cells Correlates with Suppression of Psoriasis and Colitis. Int. Immunopharmacol. 2002, 2, 653–672.
- Takahashi, H.; Kouno, M.; Nagao, K.; Wada, N.; Hata, T.; Nishimoto, S.; Iwakura, Y.; Yoshimura, A.; Yamada, T.; Kuwana, M.; et al. Desmoglein 3-Specific CD4+ T Cells Induce Pemphigus Vulgaris and Interface Dermatitis in Mice. J. Clin. Investig. 2011, 121, 3677–3688.
- Breban, M.; Fernández-Sueiro, J.L.; Richardson, J.A.; Hadavand, R.R.; Maika, S.D.; Hammer, R.E.; Taurog, J.D. T Cells, but Not Thymic Exposure to HLA-B27, Are Required for the Inflammatory Disease of HLA-B27 Transgenic Rats. J. Immunol. 1996, 156, 794–803.
- Hong, K.; Chu, A.; Lú, B.R.; Berg, E.L.; Ehrhardt, R.O. IL-12, Independently of IFN-gamma, Plays a Crucial Role in the Pathogenesis of a Murine Psoriasis-Like Skin Disorder. J. Immunol. 1999, 162, 7480–7491.
- Nishimoto, S.; Kotani, H.; Tsuruta, S.; Shimizu, N.; Ito, M.; Shichita, T.; Morita, R.; Takahashi, H.; Amagai, M.; Yoshimura, A. Th17 Cells Carrying TCR Recognizing Epidermal Autoantigen Induce Psoriasis-like Skin Inflammation. J. Immunol. 2013, 191, 3065–3072.
- Dort, E.N.; Tanguay, P.; Hamelin, R.C. CRISPR/Cas9 Gene Editing: An Unexplored Frontier for Forest Pathology. Front. Plant Sci. 2020, 11, 1126.
- Strzyz, P. CRISPR–Cas9 Wins Nobel. Nat. Rev. Mol. Cell Biol. 2020, 21, 714.
- Roth-Carter, Q.R.; Godsel, L.; Koetsier, J.L.; Broussard, J.A.; Burks, H.E.; Fitz, G.; Huffine, A.L.; Amagai, S.; Lloyd, S.; Kweon, J.; et al. 225 Desmoglein 1 Deficiency in Knockout Mice Impairs Epidermal Barrier Formation and Results in a Psoriasis-like Gene Signature in E18.5 Embryos. J. Investig. Dermatol. 2020, 140, S26.
- Godsel, L.M.; Roth-Carter, Q.R.; Koetsier, J.L.; Tsoi, L.C.; Broussard, J.A.; Fitz, G.N.; Lloyd, S.M.; Kweon, J.; Huffine, A.L.; Burks, H.E.; et al. Th17-Skewed Inflammation Due to Genetic Deficiency of a Cadherin Stress Sensor. bioRxiv 2020.
- Ippagunta, S.K.; Gangwar, R.; Finkelstein, D.; Vogel, P.; Pelletier, S.; Gingras, S.; Redeckea, V.; Häckera, H. Keratinocytes Contribute Intrinsically to Psoriasis upon Loss of TNIP1 Function. Proc. Natl. Acad. Sci. USA 2016, 113, E6162–E6171.
- Kowalczyk, T.; Ciborowski, M.; Kisluk, J.; Kretowski, A.; Barbas, C. Mass Spectrometry Based Proteomics and Metabolomics in Personalized Oncology. Biochim. Biophys. Acta (BBA)—Mol. Basis Dis. 2020, 1866, 165690.
- Rozanova, S.; Barkovits, K.; Nikolov, M.; Schmidt, C.; Urlaub, H.; Marcus, K. Quantitative Mass Spectrometry-Based Proteomics: An Overview. In Methods in Molecular Biology; Humana: New York, NY, USA, 2021; Volume 2228, pp. 85–116.
- Zhou, Y.; Wang, P.; Yan, B.X.; Chen, X.Y.; Landeck, L.; Wang, Z.Y.; Li, X.X.; Zhang, J.; Zheng, M.; Man, X.Y. Quantitative Proteomic Profile of Psoriatic Epidermis Identifies OAS2 as a Novel Biomarker for Disease Activity. Front. Immunol. 2020, 11, 1432.
- Schonthaler, H.B.; Guinea-Viniegra, J.; Wculek, S.K.; Ruppen, I.; Ximénez-Embún, P.; Guío-Carrión, A.; Navarro, R.; Hogg, N.; Ashman, K.; Wagner, E.F. S100A8-S100A9 Protein Complex Mediates Psoriasis by Regulating the Expression of Complement Factor C3. Immunity 2013, 39, 1171–1181.
- Yan, K.X.; Meng, Q.; He, H.; Zhu, H.W.; Wang, Z.C.; Han, L.; Huang, Q.; Zhang, Z.H.; Yawalkar, N.; Zhou, H.; et al. ITRAQ-Based Quantitative Proteomics Reveals Biomarkers/Pathways in Psoriasis That Can Predict the Efficacy of Methotrexate. J. Eur. Acad. Dermatol. Venereol. 2022, 36, 1784–1795.
- Li, Y.; Lin, P.; Wang, S.; Li, S.; Wang, R.; Yang, L.; Wang, H. Quantitative Analysis of Differentially Expressed Proteins in Psoriasis Vulgaris Using Tandem Mass Tags and Parallel Reaction Monitoring. Clin. Proteom. 2020, 17, 30.
- Gęgotek, A.; Domingues, P.; Wroński, A.; Wójcik, P.; Skrzydlewska, E. Proteomic Plasma Profile of Psoriatic Patients. J. Pharm. Biomed. Anal. 2018, 155, 185–193.
- Gęgotek, A.; Domingues, P.; Wroński, A.; Ambrożewicz, E.; Skrzydlewska, E. The Proteomic Profile of Keratinocytes and Lymphocytes in Psoriatic Patients. Proteom. Clin. Appl. 2019, 13, 1800119.
- Gęgotek, A.; Domingues, P.; Wroński, A.; Skrzydlewska, E. Changes in Proteome of Fibroblasts Isolated from Psoriatic Skin Lesions. Int. J. Mol. Sci. 2020, 21, 5363.
- Xu, M.; Deng, J.; Xu, K.; Zhu, T.; Han, L.; Yan, Y.; Yao, D.; Deng, H.; Wang, D.; Sun, Y.; et al. In-Depth Serum Proteomics Reveals Biomarkers of Psoriasis Severity and Response to Traditional Chinese Medicine. Theranostics 2019, 9, 2475–2488.
- Reindl, J.; Pesek, J.; Krüger, T.; Wendler, S.; Nemitz, S.; Muckova, P.; Büchler, R.; Opitz, S.; Krieg, N.; Norgauer, J.; et al. Proteomic Biomarkers for Psoriasis and Psoriasis Arthritis. J. Proteom. 2016, 140, 55–61.
- Plavina, T.; Hincapie, M.; Wakshull, E.; Subramanyam, M.; Hancock, S.W. Increased Plasma Concentrations of Cytoskeletal and Aa2+-Binding Proteins and Their Peptides in Psoriasis Patients. Clin. Chem. 2008, 54, 1805–1814.
- Matsuura, T.; Sato, M.; Nagai, K.; Sato, T.; Arito, M.; Omoteyama, K.; Suematsu, N.; Okamoto, K.; Kato, T.; Soma, Y.; et al. Serum Peptides as Putative Modulators of Inflammation in Psoriasis. J. Dermatol. Sci. 2017, 87, 36–49.
- Li, S.S.; Liu, Y.; Li, H.; Wang, L.-P.; Xue, L.-F.; Yin, G.-S.; Wu, X.-S. Identification of Psoriasis Vulgaris Biomarkers in Human Plasma by Non-Targeted Metabolomics Based on UPLC-Q-TOF/MS. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 3940–3950.
- Kishikawa, T.; Arase, N.; Tsuji, S.; Maeda, Y.; Nii, T.; Hirata, J.; Suzuki, K.; Yamamoto, K.; Masuda, T.; Ogawa, K.; et al. Large-Scale Plasma-Metabolome Analysis Identifies Potential Biomarkers of Psoriasis and Its Clinical Subtypes. J. Dermatol. Sci. 2021, 102, 78–84.
- Sun, L.; Guo, X.; Qin, Y.; Li, P.; Yu, C.; Gao, X.; Xie, X.; Xu, X. Serum Intestinal Metabolites Are Raised in Patients with Psoriasis and Metabolic Syndrome. Clin. Cosmet. Investig. Dermatol. 2022, 15, 879–886.
- Armstrong, A.W.; Wu, J.; Johnson, M.A.; Grapov, D.; Azizi, B.; Dhillon, J.; Fiehn, O. Metabolomics in Psoriatic Disease: Pilot Study Reveals Metabolite Differences in Psoriasis and Psoriatic Arthritis. F1000Research 2014, 3, 248.
- Kang, H.; Li, X.; Zhou, Q.; Quan, C.; Xue, F.; Zheng, J.; Yu, Y. Exploration of Candidate Biomarkers for Human Psoriasis Based on Gas Chromatography-Mass Spectrometry Serum Metabolomics. Br. J. Dermatol. 2017, 176, 713–722.
- Looby, N.; Roszkowska, A.; Reyes-Garcés, N.; Yu, M.; Bączek, T.; Kulasingam, V.; Pawliszyn, J.; Chandran, V. Serum Metabolic Fingerprinting of Psoriasis and Psoriatic Arthritis Patients Using Solid-Phase Microextraction—Liquid Chromatography—High-Resolution Mass Spectrometry. Metabolomics 2021, 17, 59.
- Mysliwiec, H.; Harasim-Symbor, E.; Baran, A.; Szterling-Jaworowska, M.; Milewska, A.J.; Chabowski, A.; Flisiak, I. Abnormal Serum Fatty Acid Profile in Psoriatic Arthritis. Arch. Med. Sci. 2019, 15, 1407–1414.
- Ambrożewicz, E.; Wójcik, P.; Wroński, A.; Łuczaj, W.; Jastrzab, A.; Žarković, N.; Skrzydlewska, E. Pathophysiological Alterations of Redox Signaling and Endocannabinoid System in Granulocytes and Plasma of Psoriatic Patients. Cells 2018, 7, 159.
- Tsoukalas, D.; Fragoulakis, V.; Sarandi, E.; Docea, A.O.; Papakonstaninou, E.; Tsilimidos, G.; Anamaterou, C.; Fragkiadaki, P.; Aschner, M.; Tsatsakis, A.; et al. Targeted Metabolomic Analysis of Serum Fatty Acids for the Prediction of Autoimmune Diseases. Front. Mol. Biosci. 2019, 6, 120.
- Bilgiç, Ö.; Altınyazar, H.C.; Baran, H.; Ünlü, A. Serum Homocysteine, Asymmetric Dimethyl Arginine (ADMA) and Other Arginine–NO Pathway Metabolite Levels in Patients with Psoriasis. Arch. Dermatol. Res. 2015, 307, 439–444.
- Sikora, M.; Kiss, N.; Stec, A.; Giebultowicz, J.; Samborowska, E.; Jazwiec, R.; Dadlez, M.; Olszewska, M.; Rudnicka, L. Trimethylamine N-Oxide, a Gut Microbiota-Derived Metabolite, Is Associated with Cardiovascular Risk in Psoriasis: A Cross-Sectional Pilot Study. Dermatol. Ther. 2021, 11, 1277–1289.
- Chen, C.; Hou, G.; Zeng, C.; Ren, Y.; Chen, X.; Peng, C. Metabolomic Profiling Reveals Amino Acid and Carnitine Alterations as Metabolic Signatures in Psoriasis. Theranostics 2020, 11, 754–767.
- Ramessur, R.; Corbett, M.; Marshall, D.; Acencio, M.L.; Barbosa, I.A.; Dand, N.; Di Meglio, P.; Haddad, S.; Jensen, A.H.M.; Koopmann, W.; et al. Biomarkers of Disease Progression in People with Psoriasis: A Scoping Review. Br. J. Dermatol. 2022, 187, 481–493.
- Shelef, M.A.; Sokolove, J.; Lahey, L.J.; Wagner, C.A.; Sackmann, E.K.; Warner, T.F.; Wang, Y.; Beebe, D.J.; Robinson, W.H.; Huttenlocher, A. Peptidylarginine Deiminase 4 Contributes to Tumor Necrosis Factor α-Induced Inflammatory Arthritis. Arthritis Rheumatol. 2014, 66, 1482–1491.
- Kiss, B.; Szántó, M.; Hegedűs, C.; Antal, D.; Szödényi, A.; Márton, J.; Méhes, G.; Virág, L.; Szegedi, A.; Bai, P. Poly(ADP-Ribose) Polymerase-1 Depletion Enhances the Severity of Inflammation in an Imiquimod-Induced Model of Psoriasis. Exp. Dermatol. 2020, 29, 79–85.
- Sestito, R.; Madonna, S.; Scarponi, C.; Cianfarani, F.; Failla, C.M.; Cavani, A.; Girolomoni, G.; Albanesi, C. STAT3-dependent Effects of IL-22 in Human Keratinocytes Are Counterregulated by Sirtuin 1 through a Direct Inhibition of STAT3 Acetylation. FASEB J. 2011, 25, 916–927.
- Fan, X.; Yan, K.; Meng, Q.; Sun, R.; Yang, X.; Yuan, D.; Li, F.; Deng, H. Abnormal Expression of SIRTs in Psoriasis: Decreased Expression of SIRT 1-5 and Increased Expression of SIRT 6 and 7. Int. J. Mol. Med. 2019, 44, 157–171.
- Bohio, A.A.; Sattout, A.; Wang, R.; Wang, K.; Sah, R.K.; Guo, X.; Zeng, X.; Ke, Y.; Boldogh, I.; Ba, X. C-Abl–Mediated Tyrosine Phosphorylation of PARP1 Is Crucial for Expression of Proinflammatory Genes. J. Immunol. 2019, 203, 1521–1531.
- Łuczaj, W.; Gęgotek, A.; Skrzydlewska, E. Analytical Approaches to Assess Metabolic Changes in Psoriasis. J. Pharm. Biomed. Anal. 2021, 205, 114359.
- Lundberg, K.C.; Fritz, Y.; Johnston, A.; Foster, A.M.; Baliwag, J.; Gudjonsson, J.E.; Schlatzer, D.; Gokulrangan, G.; McCormick, T.S.; Chance, M.R.; et al. Proteomics of Skin Proteins in Psoriasis: From Discovery and Verification in a Mouse Model to Confirmation in Humans. Mol. Cell. Proteom. 2015, 14, 109–119.
- Boehncke, W.H. Systemic Inflammation and Cardiovascular Comorbidity in Psoriasis Patients: Causes and Consequences. Front. Immunol. 2018, 9, 579.




