
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Luisa Ladel | -- | 1850 | 2023-08-04 20:49:51 | | | |
| 2 | Alfred Zheng | Meta information modification | 1850 | 2023-08-08 02:35:41 | | | | |
| 3 | Alfred Zheng | -2 word(s) | 1848 | 2023-09-05 11:25:39 | | |
Video Upload Options
Appendiceal cancers (AC) are a rare and heterogeneous group of malignancies. Historically, appendiceal neoplasms have been grouped with colorectal cancers (CRC), and treatment strategies have been modeled after CRC management guidelines due to their structural similarities and anatomical proximity.
1. Introduction
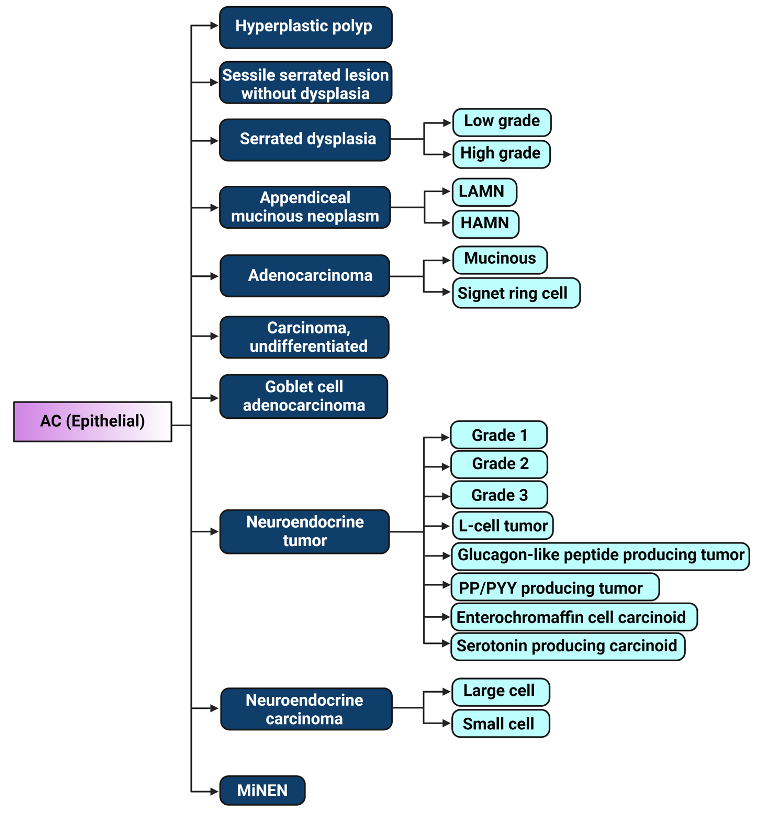
Figure 1. Illustration of 2019 WHO classification of epithelial appendiceal cancers [23]. LAMN: low-grade appendiceal mucinous neoplasm, HAMN: high-grade appendiceal mucinous neoplasm, PP/PYY: pancreatic polypeptide/peptide YY, MiNEN: Mixed neuroendocrine – non-neuroendocrine neoplasm. Figure created with biorender.com.
2. Chromatin Remodeling and Transcription in Appendiceal Cancers
Epigenetics modifications play an important role in chromatin modulation and are subject to environmental forces typically through its dynamic and reversible nature. However, they can also be heritable and persist over several generations [17]. The major epigenetic mechanisms include methylation, leading to the suppression or silencing of gene activation, and acetylation, causing the activation of transcription, both of which can take place on histones, affecting large areas of the genome, or in a more specific manner on DNA itself at the CpG sites of the promoter regions of specific genes. The other main categories of epigenetic mechanisms include chromatin remodeling by nucleosome positioning and regulation via non-coding RNAs [18]. Epigenetic changes in malignancy have attracted much attention, especially in gastrointestinal neoplasms, since they often occur early in carcinogenesis and involve key cancer-associated pathways [19][20]. Burgeoning evidence has shown that epigenetic signatures constitute crucial hallmarks of disease pathogenesis. This field has become an area of intensive research for biomarker development and novel therapeutic strategies in the era of precision medicine [20]. To our knowledge, appendiceal cancers have no established epigenetic alterations or signatures. However, mutational genomic data and pathway enrichment analysis from several molecular studies of appendiceal cancers has revealed genes and pathways that could potentially be involved in epigenetic regulation. The exploration of these genes and their regulatory pathways could provide deeper insights into the epigenetic landscape of appendiceal cancers.
2.1. SWI/SNF Chromatin Remodeling Complex
2.2. COMPASS Chromatin Remodeling Complex
2.3. The Forkhead Box O (FoxO) Transcription Factors
A tremendous breakthrough in medical oncology was achieved with the introduction of immune checkpoint inhibitors. Exciting data proposes potential for treatment synergism between immunotherapy and epigenetic drugs, such as DNA demethylating agents. It has been shown that treatment with this class of drugs creates an Interferon-mediated immune response within the tumor microenvironment of hematological, ovarian, and colorectal cancers [38][39][40]. This is thought to enhance the efficiency of the antitumoral immune response, which has been hypothesized to increase even further in combination with immune checkpoint blockers. Furthermore, dysregulation of epigenetic silencing by DNMT1 inhibition via PI3K/AKT hyperactivation and aberrant activation of the TGFβ signaling pathway have been unmasked as key drivers behind immune evasion and lack of response to immunotherapy [41]. Other studies have revealed enhanced sensitivity to immune checkpoint blockade in tumors carrying SWI/SNF complex mutations. ARID1A deficiency led to significantly reduced tumor burden and prolonged survival upon immunotherapy compared to wild-type tumors in studies for ovarian and gastric cancers [21]. Likewise, deviant transcriptional regulation due to inhibition of EZH2 has been implicated in correlating with the immunogenicity of tumor cells and immune silencing in the tumor microenvironment. The utility of combination therapies with EZH2 inhibitors and immune checkpoint blockers remains to be investigated further, with several clinical trials underway [42].
Exciting advances have also been made in recent years in EZH2-targeted therapies. EZH2 is mutated in specific forms of appendiceal cancers, and several of the other epigenetic regulators found to be mutated in appendiceal neoplasms have been linked in some form to EZH2 overexpression or hyperactivation as well; most prominently, PI3K/AKT, as well as KDM6A and specific subunits of the SWI/SNF complex. This makes EZH2 a prime therapeutic target, and several compounds have been developed since EZH2 inhibitor Tazemetostat was FDA approved for advanced epitheloid sarcoma as well as relapsed or refractory follicular lymphoma, with several ongoing phase 1 and 2 clinical trials investigating similar drugs, such as SHR2554 and CPI-1205 (or lirametostat) in small intestine neuroendocrine tumors and relapsed or refractory B-cell/T-cell and Hodgkin’s lymphomas, respectively [43][44][45]. Another study linked EZH2-mediated epigenetic changes to chromatin density to increased resistance to DNA damage in cells with concurrent p53 mutation and presented data suggesting that resistance to treatment approaches with chemotherapy and radiation as conferred by p53 mutation could be overcome, at least in part, by EZH2 inhibition [42]. However, more than direct targeting of these mutations or their affected pathways are attainable treatment approach. There are, for example, encouraging data proposing the utility of existing DNA damage repair inhibitors in tumors with KMT2D mutations. These findings align with the increased susceptibility to DNA damage found in KMT2D-deficient tumors, as evidenced by increased sensitivity to PARP inhibitors [31]. Similar findings were obtained in tumors with mutations affecting the SWI/SNF complex. Specifically, PARP inhibitors are under investigation in several trials for tumors with ARID1A mutation, based on the involvement of the SWI/SNF complex in DNA damage repair and therapeutic vulnerability observed in preclinical studies [21].

Figure 2. Major epigenetic pathways contributing to oncogenesis in appendiceal cancers and potential therapy-related targets. Ac: Acetylation, DNMT: DNA Methyltransferase, KAc: Lysine acetylation, Me: Methylation, P: Phosphorylation, RTK: Receptor Tyrosine Kinase, TSS: Transcription start site, Ub: Ubiquitination. Figure created with biorender.com
3. Conclusions
The intricacies of epigenetic alterations and mechanisms in appendiceal neoplasms are still largely unknown. However, several epigenetic mechanisms have been postulated based on currently available data, which hold highly promising potential for clinical applicability regarding novel diagnostics and prognostication in appendiceal cancers. Further studies are necessary to validate previous findings in a methodical, epigenomics-centered, and translational approach. Epigenetics-based biomarkers may be the key to a deeper understanding of epithelial appendiceal cancer pathophysiology and aid in uncovering actionable targets for disease monitoring in appendiceal cancers. Ultimately this could enable clinicians to prognosticate responses to various therapy approaches, estimate the risk of progression or relapse and predict overall survival in their patients, thereby making personalized oncology a reality in managing and treating appendiceal neoplasms.
References
- Salazar, M.C.; Canavan, M.E.; Chilakamarry, S.; Boffa, D.J.; Schuster, K.M. Appendiceal Cancer in the National Cancer Database: Increasing Frequency, Decreasing Age, and Shifting Histology. J. Am. Coll. Surg. 2022, 234, 1082–1089.
- Marmor, S.; Portschy, P.R.; Tuttle, T.M.; Virnig, B.A. The rise in appendiceal cancer incidence: 2000–2009. J. Gastrointest. Surg. 2015, 19, 743–750.
- Yilmaz, S.; Bolukbasi, H. Appendiceal neoplasms: Suspected findings and reports of 14 cases. Indian J. Cancer 2022.
- O’Donnell, M.E.; Badger, S.A.; Beattie, G.C.; Carson, J.; Garstin, W.I. Malignant neoplasms of the appendix. Int. J. Colorectal. Dis. 2007, 22, 1239–1248.
- McGory, M.L.; Maggard, M.A.; Kang, H.; O’Connell, J.B.; Ko, C.Y. Malignancies of the appendix: Beyond case series reports. Dis. Colon Rectum 2005, 48, 2264–2271.
- McCusker, M.E.; Coté, T.R.; Clegg, L.X.; Sobin, L.H. Primary malignant neoplasms of the appendix: A population-based study from the surveillance, epidemiology and end-results program, 1973–1998. Cancer 2002, 94, 3307–3312.
- Hatch, Q.M.; Gilbert, E.W. Appendiceal Neoplasms. Clin. Colon Rectal Surg. 2018, 31, 278–287.
- American Cancer Society, National Cancer Institute, and the National Organization for Rare Disorders. Appendix Cancer: Statistics. 2022. Available online: https://www.cancer.net/cancer-types/appendix-cancer/statistics (accessed on 30 March 2023).
- Alajääski, J.; Lietzén, E.; Grönroos, J.M.; Mecklin, J.P.; Leppäniemi, A.; Nordström, P.; Rautio, T.; Rantanen, T.; Sand, J.; Paajanen, H.; et al. The association between appendicitis severity and patient age with appendiceal neoplasm histology-a population-based study. Int. J. Colorectal. Dis. 2022, 37, 1173–1180.
- Skendelas, J.P.; Alemany, V.S.; Au, V.; Rao, D.; McNelis, J.; Kim, P.K. Appendiceal adenocarcinoma found by surgery for acute appendicitis is associated with older age. BMC Surg. 2021, 21, 228.
- van den Heuvel, M.G.; Lemmens, V.E.; Verhoeven, R.H.; de Hingh, I.H. The incidence of mucinous appendiceal malignancies: A population-based study. Int. J. Colorectal. Dis. 2013, 28, 1307–1310.
- Singh, H.; Koomson, A.S.; Decker, K.M.; Park, J.; Demers, A.A. Continued increasing incidence of malignant appendiceal tumors in Canada and the United States: A population-based study. Cancer 2020, 126, 2206–2216.
- Van de Moortele, M.; De Hertogh, G.; Sagaert, X.; Van Cutsem, E. Appendiceal cancer: A review of the literature. Acta Gastroenterol. Belg. 2020, 83, 441–448.
- Flum, D.R.; Davidson, G.H.; Monsell, S.E.; Shapiro, N.I.; Odom, S.R.; Sanchez, S.E.; Drake, F.T.; Fischkoff, K.; Johnson, J.; Patton, J.H.; et al. A Randomized Trial Comparing Antibiotics with Appendectomy for Appendicitis. N. Engl. J. Med. 2020, 383, 1907–1919.
- Sallinen, V.; Akl, E.A.; You, J.J.; Agarwal, A.; Shoucair, S.; Vandvik, P.O.; Agoritsas, T.; Heels-Ansdell, D.; Guyatt, G.H.; Tikkinen, K.A. Meta-analysis of antibiotics versus appendicectomy for non-perforated acute appendicitis. Br. J. Surg. 2016, 103, 656–667.
- Newdow, M. Management of Acute Appendicitis—Longer-Term Outcomes. N. Engl. J. Med. 2022, 386, 900.
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341.
- Fardi, M.; Solali, S.; Farshdousti Hagh, M. Epigenetic mechanisms as a new approach in cancer treatment: An updated review. Genes Dis. 2018, 5, 304–311.
- Sharma, S.; Kelly, T.K.; Jones, P.A. Epigenetics in cancer. Carcinogenesis 2010, 31, 27–36.
- Cheng, Y.; He, C.; Wang, M.; Ma, X.; Mo, F.; Yang, S.; Han, J.; Wei, X. Targeting epigenetic regulators for cancer therapy: Mechanisms and advances in clinical trials. Signal Transduct. Target. Ther. 2019, 4, 62.
- Mittal, P.; Roberts, C.W.M. The SWI/SNF complex in cancer—Biology, biomarkers and therapy. Nat. Rev. Clin. Oncol. 2020, 17, 435–448.
- Jones, C.A.; Tansey, W.P.; Weissmiller, A.M. Emerging Themes in Mechanisms of Tumorigenesis by SWI/SNF Subunit Mutation. Epigenet. Insights 2022, 15, 25168657221115656.
- Li, Z.; Zhao, J.; Tang, Y. Advances in the role of SWI/SNF complexes in tumours. J. Cell. Mol. Med. 2023, 27, 1023–1031.
- Qi, W.; Wang, R.; Chen, H.; Wang, X.; Xiao, T.; Boldogh, I.; Ba, X.; Han, L.; Zeng, X. BRG1 promotes the repair of DNA double-strand breaks by facilitating the replacement of RPA with RAD51. J. Cell Sci. 2015, 128, 317–330.
- Watanabe, R.; Ui, A.; Kanno, S.; Ogiwara, H.; Nagase, T.; Kohno, T.; Yasui, A. SWI/SNF factors required for cellular resistance to DNA damage include ARID1A and ARID1B and show interdependent protein stability. Cancer Res. 2014, 74, 2465–2475.
- Wen, K.W.; Grenert, J.P.; Joseph, N.M.; Shafizadeh, N.; Huang, A.; Hosseini, M.; Kakar, S. Genomic profile of appendiceal goblet cell carcinoid is distinct compared to appendiceal neuroendocrine tumor and conventional adenocarcinoma. Hum. Pathol. 2018, 77, 166–174.
- Garland-Kledzik, M.; Scholer, A.; Ensenyat-Mendez, M.; Orozco, J.I.J.; Khader, A.; Santamaria-Barria, J.; Fischer, T.; Pigazzi, A.; Marzese, D.M. Establishing Novel Molecular Subtypes of Appendiceal Cancer. Ann. Surg. Oncol. 2022, 29, 2118–2125.
- Seton-Rogers, S. Pancreatic cancer: The COMPASS shows the way. Nat. Rev. Cancer 2018, 18, 373.
- Revia, S.; Seretny, A.; Wendler, L.; Banito, A.; Eckert, C.; Breuer, K.; Mayakonda, A.; Lutsik, P.; Evert, M.; Ribback, S.; et al. Histone H3K27 demethylase KDM6A is an epigenetic gatekeeper of mTORC1 signalling in cancer. Gut 2022, 71, 1613–1628.
- Lavery, W.J.; Barski, A.; Wiley, S.; Schorry, E.K.; Lindsley, A.W. KMT2C/D COMPASS complex-associated diseases : An emerging class of congenital regulopathies. Clin. Epigenet. 2020, 12, 10.
- Lv, S.; Wen, H.; Shan, X.; Li, J.; Wu, Y.; Yu, X.; Huang, W.; Wei, Q. Loss of KMT2D induces prostate cancer ROS-mediated DNA damage by suppressing the enhancer activity and DNA binding of antioxidant transcription factor FOXO3. Epigenetics 2019, 14, 1194–1208.
- Schulz, W.A.; Lang, A.; Koch, J.; Greife, A. The histone demethylase UTX/KDM6A in cancer: Progress and puzzles. Int. J. Cancer 2019, 145, 614–620.
- Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019, 28, 1947–1951.
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Kawashima, M.; Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023, 51, D587–D592.
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30.
- Yadav, R.K.; Chauhan, A.S.; Zhuang, L.; Gan, B. FoxO transcription factors in cancer metabolism. Semin. Cancer Biol. 2018, 50, 65–76.
- Lin, D.L.; Wang, L.L.; Zhao, P.; Ran, W.W.; Wang, W.; Zhang, L.X.; Han, M.; Bao, H.; Liu, K.; Wu, X.; et al. Gastrointestinal Goblet Cell Adenocarcinomas Harbor Distinctive Clinicopathological, Immune, and Genomic Landscape. Front. Oncol. 2021, 11, 758643.
- Berger, E.R.; Park, T.; Saridakis, A.; Golshan, M.; Greenup, R.A.; Ahuja, N. Immunotherapy Treatment for Triple Negative Breast Cancer. Pharmaceuticals 2021, 14, 763.
- Chiappinelli, K.B.; Zahnow, C.A.; Ahuja, N.; Baylin, S.B. Combining Epigenetic and Immunotherapy to Combat Cancer. Cancer Res. 2016, 76, 1683–1689.
- Soares, K.C.; Zheng, L.; Ahuja, N. Overcoming immune system evasion by personalized immunotherapy.. Pers. Med. 2014, 11, 561–564.
- Villanueva, L.; Álvarez-Errico, D.; Esteller, M. The Contribution of Epigenetics to Cancer Immunotherapy. Trends Immunol. 2020, 41, 676–691.
- Eich, M.L.; Athar, M.; Ferguson, J.E., 3rd; Varambally, S. EZH2-Targeted Therapies in Cancer: Hype or a Reality. Cancer Res. 2020, 80, 5449–5458.
- Gulati, N.; Béguelin, W.; Giulino-Roth, L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk. Lymphoma 2018, 59, 1574–1585.
- Straining, R.; Eighmy, W. T J. azemetostat: EZH2 Inhibitor. Adv. Pract. Oncol. 2022, 13, 158–163.
- Song, Y.; Liu, Y.; Li, Z.M.; Li, L.; Su, H.; Jin, Z.; Zuo, X.; Wu, J.; Zhou, H.; Li, K.; et al.et al. SHR2554, an EZH2 inhibitor, in relapsed or refractory mature lymphoid neoplasms: A first-in-human, dose-escalation, dose-expansion, and clinical expansion phase 1 trial. Lancet Haematol. 2022, 9, e493–e503.




