Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Francisco Javier Marcos-Torres | -- | 3185 | 2023-08-03 16:52:49 | | | |
| 2 | Conner Chen | Meta information modification | 3185 | 2023-08-07 04:59:32 | | | | |
| 3 | Conner Chen | -5 word(s) | 3180 | 2023-09-05 10:55:17 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Sánchez-Jiménez, A.; Llamas, M.A.; Marcos-Torres, F.J. Control of Pseudomonas aeruginosa Virulence by One-Component Systems. Encyclopedia. Available online: https://encyclopedia.pub/entry/47630 (accessed on 08 February 2026).
Sánchez-Jiménez A, Llamas MA, Marcos-Torres FJ. Control of Pseudomonas aeruginosa Virulence by One-Component Systems. Encyclopedia. Available at: https://encyclopedia.pub/entry/47630. Accessed February 08, 2026.
Sánchez-Jiménez, Ana, María A. Llamas, Francisco Javier Marcos-Torres. "Control of Pseudomonas aeruginosa Virulence by One-Component Systems" Encyclopedia, https://encyclopedia.pub/entry/47630 (accessed February 08, 2026).
Sánchez-Jiménez, A., Llamas, M.A., & Marcos-Torres, F.J. (2023, August 03). Control of Pseudomonas aeruginosa Virulence by One-Component Systems. In Encyclopedia. https://encyclopedia.pub/entry/47630
Sánchez-Jiménez, Ana, et al. "Control of Pseudomonas aeruginosa Virulence by One-Component Systems." Encyclopedia. Web. 03 August, 2023.
Copy Citation
Pseudomonas aeruginosa is a pathogen capable of colonizing virtually every human tissue. The host colonization competence and versatility of this pathogen are powered by a wide array of virulence factors necessary in different steps of the infection process. This includes factors involved in bacterial motility and attachment, biofilm formation, the production and secretion of extracellular invasive enzymes and exotoxins, the production of toxic secondary metabolites, and the acquisition of iron. Expression of these virulence factors during infection is tightly regulated, which allows their production only when they are needed. This process optimizes host colonization and virulence.
Pseudomonas aeruginosa
pathogenesis
virulence
1. Control of P. aeruginosa Virulence by One-Component Systems
One-component systems (OCSs) are the simplest and most responsive transcriptional regulators of bacteria. Canonical OCSs are composed of a single cytoplasmic protein that has an input or sensor domain for signal recognition and an output DNA-binding domain that responds to the signal by modifying gene transcription (Figure 1a) [1]. One-component proteins have a wide diversity of input domains that allow the detection of multiple different signals. For the output domain, around 84% of one-component proteins have a helix-turn-helix (HTH) domain, which binds to DNA and activates or represses gene transcription [1]. However, a minority of these proteins have another type of output domain including enzymatic domains that regulate cyclic nucleotides production and protein phosphorylation. The cytosolic location of one-component proteins reduces the type of external signals that can access this bacterial compartment to be recognized by the sensor domain. For this reason, one-component systems recognize signals produced in the cytosol or small molecules able to diffuse through the membrane, like light or gases [1]. Upon signal recognition, the output domain undergoes a conformational change that induces a response, usually by exposing the HTH domain of the protein and allowing its binding to DNA [2]. The presence of a sensor and effector domain in a single protein allows for a quick response to the signal in detriment to the fine-tuned and more precise response carried out by transcriptional regulators, in which these functions reside in different proteins (e.g., two-component systems). P. aeruginosa harbors between 408 and 452 OCS proteins, with the type strain PAO1 encoding 423 regulators of this type [3].
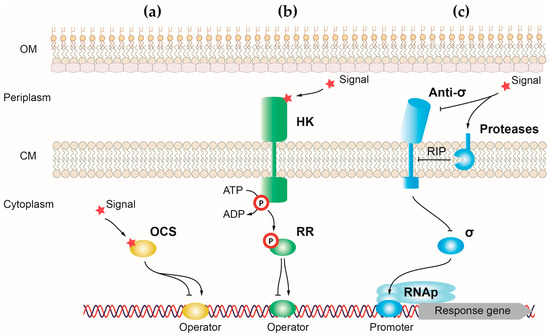
Figure 1. Schematic representation showing the three main classes of transcriptional regulators in P. aeruginosa. Mechanisms underlying gene expression regulation by (a) one-component systems, (b) two-component systems, and (c) alternative σ factors. Positive and negative controls are represented with arrows and T-shaped lines, respectively. OM, outer membrane; CM, cytoplasmic membrane; RIP, regulated intramembrane proteolysis.
2. OCSs Responding to Quorum Sensing (QS)
QS responds to cell population density, enabling bacteria to restrict the expression of specific genes to high cell densities at which the resulting phenotypes will be most beneficial. In P. aeruginosa, QS is performed by three autoinducers, two N-acyl homoserine lactones (AHLs), and a quinolone signal-designed PQS (from Pseudomonas quinolone signal) [4]. These molecules are produced by P. aeruginosa and diffuse freely through the membrane into the extracellular medium. When the cell density is high and the concentration of the autoinducer is above the threshold, it is recognized by a OCS protein that responds by modifying gene expression [4].
2.1. LasR
LasR belongs to the LuxR family of regulators, which typically acts as transcriptional activators, and is the main regulator of the Las QS system in P. aeruginosa. LasR senses the autoinducer N-(3-oxododecanoyl)homoserine lactone (3O-C12-HSL), a product of the lasI gene, and functions as a global regulator that directly or indirectly activates the expression of over 300 genes, many of which are involved in virulence [5] (Figure 2). In addition to lasI, these include the elastase genes lasA and lasB, the hcnABC cluster for the production of hydrogen cyanide, plcB phospholipase, the flp-type IVb pili subunit, aprA alkaline protease, the pyoverdine regulator pvdS, and the toxA exotoxin genes [5][6]. LasR also induces the expression of the RsaL repressor, which dampens the expression of LasR targets [7]. LasR occupies the highest position in the hierarchical QS signaling network, playing a key role activating the transcription of the Rhl and Pqs QS systems (Figure 2) [8]. Importantly, P. aeruginosa lasR and lasI mutants are considerably less virulent than the wild-type strain in a burn mouse model of infection and a rat model of acute pneumonia, and the lack of LasR produces a lower inflammatory response and impaired host colonization [8][9][10].
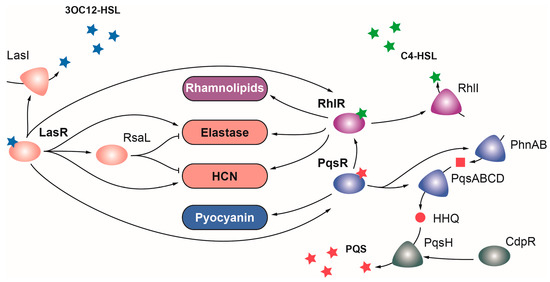
Figure 2. Regulation of virulence by quorum sensing in P. aeruginosa. The Las (depicted in pink), Rhl (depicted in purple), and PQS (depicted in blue) QS systems are governed by the OCS LasR, RhlR, and PqsR, respectively. Upon detection of their signal molecules, each OCS activates the expression of virulence factors and enzymes producing their respective inducing signals. Positive and negative regulations are represented with arrows and T-shaped lines, respectively.
2.2. RhlR
Mirroring LasR, the LuxR family regulator RhlR is the transcriptional activator of the Rhl QS system, which detects the N-butanoyl-L-homoserine lactone (C4-HSL) signal molecule generated by the product of the rhlI gene [11]. RhlR directly or indirectly activates the expression of over 100 genes, including several virulence genes like rhlA and rhlB for synthesis of rhamnolipids, and LasR targets lasB, hcnABC, and phenazine genes [8] (Figure 2). P. aeruginosa rhlR and rhlI mutants are considerably less virulent than the wild-type strain in murine infection models [12][13]. Despite the similarities and overlapping functions between the Las and Rhl systems, each one contributes to virulence independently, as evidenced by the synergistic effects on virulence observed when both systems are mutated [6][14]. A double lasR rhlR mutant resulted in lower mortality rates for the nematode C. elegans than single lasR and rhlR mutants [14]. Similarly, a lasI rhlI double mutant was more lethal than single mutants in a burn mouse model of infection [6]. Surprisingly, whilea lasR or rhlR single mutation reduces the cytotoxicity of P. aeruginosa towards lung epithelial cells, the cytotoxicity of the lasR rhlR double mutant is higher than that of the wild-type strain [14]. This suggests that some elements of the LasR and RhlR regulons may act in opposite directions [14].
2.3. PqsR
The third QS regulator of P. aeruginosa is PqsR (also named MvfR), a transcriptional regulator of the LysR family that senses PQS, a quinole synthesized by the pqsABCDE and phnAB operons and the pqsH gene [15]. PqsR upregulates the expression of more than 100 genes, including the pqsABCDE and phnAB operons, and many genes of the RhlR regulon [15] (Figure 2). This overlap between these two QS systems seems to arise from PqsE thioesterase, which is not only required for PQS synthesis but also for RhlR activity [15]. Among others, PqsR activates the expression of genes encoding the virulence factors pyocyanin and hydrogen cyanide and the lectin LecA [15]. A pqsR mutation reduces the mortality rate caused by P. aeruginosa in a burn mouse model of infection, but this effect is likely due to reduced expression of the pqsE gene, and therefore to the reduced RhlR activity in this mutant [15].
Moreover, PQS regulates the expression of many genes involved in iron acquisition such as pyoverdine and pyochelin biosynthetic genes [16]. This is likely an indirect effect due to the iron-chelating properties of PQS [16][17]. Interestingly, PqsR not only responds to PQS but also to its precursor molecule HHQ [8]. The pqsH gene encoding the enzyme required for the conversion of HHQ into PQS is the only gene in the biosynthetic cluster that is not regulated by PqsR but by the AraC family regulator CdpR [8][18]. According to the repressor role of this regulator, a P. aeruginosa cdpR mutant produced a higher mortality rate and increased lung injury and inflammation in a mouse model of acute pneumonia [18].
3. OCSs Regulating Motility, Attachment, and Biofilm Formation
AmrZ
The P. aeruginosa AmrZ (from alginate and motility regulator) belongs to the ribbon-helix-helix family of transcriptional regulators, which mainly functions as repressors [19]. While acting as a repressor for nearly 50 genes, AmrZ can also act as an activator, controlling the expression of virulence genes such as P. aeruginosa alginate synthesis and transport genes encoded within the alg cluster [20][21] (Figure 3). Because alginate production promotes P. aeruginosa biofilm formation and persistence, by activating the expression of the alg operon, AmrZ promotes this virulence phenotype. However, in response to changes in oxygen and nutrient levels or in the presence of toxic elements such as nitric oxide, AmrZ promotes cell dispersion from bacterial biofilms by activating the expression of several hydrolase and nuclease genes such as endA, pelA, and pslG [22]. AmrZ also plays a major role in motility, acting as a repressor of the type IV pili precursor pilA and fleQ, a c-di-GMP effector that activates the transcription of flagellum synthesis genes [23].
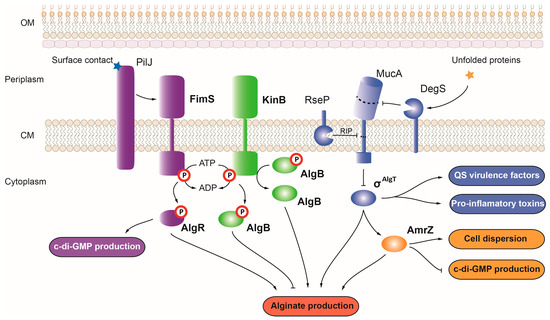
Figure 3. Regulation of alginate production and virulence in P. aeruginosa. Alginate production in P. aeruginosa is controlled by the OCS AmrZ (depicted in yellow), the TCSs FimS-AlgR and KinB-AlgB (depicted in purple and green, respectively), and the σAlgT factor (depicted in blue). Positive and negative controls are represented with arrows and T-shaped lines, respectively. OM, outer membrane; CM, cytoplasmic membrane; RIP, regulated intramembrane proteolysis.
4. OCSs Regulating the Production and Secretion of Extracellular Enzymes, Toxins, and Toxic Secondary Metabolites
4.1. ExsA
ExsA belongs to the AraC family of regulators that mostly act as transcriptional activators upon signal sensing [24]. This regulator induces the expression of P. aeruginosa T3SS in response to low calcium levels, recruiting the RNAP complex to the promoter of the T3SS cluster [25]. This results in transcription from the 11 promoters controlling T3SS expression and secretion of the ExoT, ExoS, ExoU, and ExoY effectors. ExsA was recently found to also activate the transcription of the impA gene that specifies a metalloprotease secreted by T2SS, which confers resistance to phagocytosis from macrophages [26]. In contrast to most AraC regulators that are activated by a small-molecule ligand, ExsA is part of the ExsACDE signaling cascade [25]. In non-induced conditions, ExsA is sequestered by ExsD, and the chaperone ExsC forms a complex with ExsE. Contact with host cells causes T3SS to secrete ExsE, switching ExsC to pair with ExsD instead. ExsA, now free from ExsD repression, activates gene expression [25]. Cytotoxicity assays showed that the cytotoxicity of a P. aeruginosa exsA mutant toward different cell lines was severely impaired [27][28]. Moreover, an exsA mutant is more susceptible to phagocytosis than the wild-type strain, likely due to the ExsA-dependent transcription of impA [26][29].
4.2. Sfa2
The Sfa2 protein is an EBP that activates the σ54-dependent transcription of P. aeruginosa H2-T6SS. Sfa2 binds to the enhancer-binding site on H2-T6SS genes, promoting the transcription of ~15 genes in this gene cluster, which includes the sfa2 gene itself [30]. A paralogue EBP regulator known as Sfa3 is encoded within the P. aeruginosa H3-T6SS gene cluster, suggesting a similar control of H3-T6SS expression by σ54 and this EBP [31].
4.3. GbdR
The AraC regulator GbdR activates the expression of hemolytic phospholipase C PlcH upon sensing glycine betaine and dimethylglycine [32]. PlcH produces phosphorylcholine, which is then dephosphorylated by PchP to release choline. Choline metabolism leads to the production of glycine betaine, which can either accumulate as an osmoprotector and a bacterial surfactant or be used as a nutrient source. GbdR acts as a switch for the transition between both fates upon accumulation of glycine betaine by promoting the expression of genes involved in choline conversion into serine and pyruvate [32]. Expression of the gbdR gene is activated by σRpoN and the EBPs CbrB and NtrC in the absence of carbon/nitrogen sources, and it is repressed by the betBA cluster repressor BetI in the absence of choline [33]. Although the role of GbdR in virulence has not been assayed to date, choline metabolism was shown to be important for the proper colonization and survival of P. aeruginosa in a mouse model of lung infection [34].
4.4. PvrA
The Pseudomonas virulence regulator PvrA belongs to the TetR family of transcriptional regulators that normally act as repressors of gene expression. Like GbdR, PvrA promotes the expression of the plcH phospholipase gene, although in response to fatty acyl-CoAs such as palmitoyl-CoA [35][36]. PvrA serves as a link between virulence and metabolism of phosphatidylcholine and long-chain fatty acids, one of the main carbon sources for P. aeruginosa during lung infection and a major component of the lung surfactant. In addition to directly controlling the expression of several metabolism genes and aprA alkaline protease, PvrA activates the production of pyocyanin and rhamnolipids by promoting transcription of the PQS biosynthesis operons of pqsABCDE and phnAB [35][36]. Moreover, it represses the expression of phaG encoding a protein involved in the conversion of 3-hydroxyacyl-acyl carrier protein (ACP), the substrate required for rhamnolipids production, into polyhydroxyalkanoate energy storage compounds [35]. In this way, PvrA acts as a switch diverting 3-hydroxyacyl-ACP use toward rhamnolipids production instead of energy storage. Accordingly, a mutation of pvrA was shown to significantly reduce the colonization and survival of P. aeruginosa in the lungs of mice [35][36].
4.5. SphR
SphR is an AraC family regulator that responds to sphingosine and activates the expression of genes that P. aeruginosa uses to feed on sphingolipids from the host cell membranes and the surfactant of mammalian lungs [37][38]. SphR activates the expression of the sphingolipid metabolism genes sphA and sphBCD and the ceramidase gene cerN [37][38]. In addition to enhancing the activity of PlcH phospholipase, CerN is involved in the conversion of ceramide into sphingosine and fatty acids. P. aeruginosa uses fatty acids as a nutrient source, while sphingosine is used to attack host cells by disrupting the skin barrier [37]. SphR was found to be important for P. aeruginosa survival inside the lungs of mice, especially to overcome the antimicrobial effects of sphingosine [37][38].
4.6. PtxR
PtxR belongs to the LysR family of regulators, generally regarded as transcriptional activators of their target genes but with repressor activity on their own genes [39]. PtxR activates the transcription of the exotoxin A gene toxA and the lasB elastase and represses the transcription of rhlA rhamnolipids and phzA1 phenazine genes [40]. The activity of PtxR is modulated by the 2-ketogluconate sensor PtxS, which always acts as a repressor of gene expression regardless of PtxR’s role [41]. When PtxR acts as a repressor, PtxS binds to both DNA and PtxR, creating a DNA loop that promotes repressor activity. Repression is relieved by the presence of 2-ketogluconate, which releases both proteins from DNA. Conversely, when PtxR acts as an activator, PtxS binds to PtxR–DNA complexes, preventing gene expression. Upon detection of 2-ketogluconate, PtxS dissociates from PtxR, which activates gene expression [41][42].
4.7. SoxR
SoxR belongs to the MerR family of transcriptional regulators, which typically act as activators of gene expression [43]. SoxR regulators normally respond to oxidative stress by sensing superoxide; however, in P. aeruginosa, SoxR is activated by pyocyanin in a superoxide-independent manner and controls the release of phenazines and other chorismate-derived compounds [44][45]. To date, only two gene clusters have been identified as being part of the SoxR regulon: monooxygenase PA2274 and the multidrug efflux pump operon mexGHI-opmD [44][46]. The efflux pump MexGHI-OpmD was shown to increase the resistance to antibiotics such as norfloxacin and tetracycline, and it is involved in the export of a phenazine precursor required for pyocyanin production [47][48]. This efflux pump is also required for the production of virulence factors such as LasB elastase, rhamnolipids, pyocyanine, and pyoverdine, probably caused by the genetic response triggered by phenazines as signal molecules [47]. Accordingly, a P. aeruginosa soxR mutant was found to be attenuated for virulence in pulmonary and burn mouse models of infection [46][49].
5. OCSs Regulating the Acquisition and Homeostasis of Iron
5.1. Fur
The ferric uptake regulator Fur is the main regulator of iron homeostasis in P. aeruginosa. This regulator facilitates host infection by maintaining sufficient intracellular iron levels while avoiding iron toxicity. Upon binding iron, Fur directly represses the expression of iron acquisition genes and indirectly activates the expression of iron storage and detoxification genes, like the bacterioferritin gene bfrB, superoxide dismutase sodB, and catalase katA [17][50]. Fur expands its regulation of iron homeostasis by repressing the expression of two sRNAs, i.e., PrrF1 and PrrF2, which block the production of over 50 proteins by binding their mRNA targets [17][51]. Fur represses the expression of pyoverdine and pyochelin biosynthesis and transport genes, which are thus only produced under iron-limited conditions [17]. Expression of the hemophore HasAp and the Phu, Has, and Hxu heme transport systems is also repressed by Fur under iron sufficient conditions. Moreover, Fur also represses the expression of other virulence factors including exotoxin A, the PIV/PrpL and IcmP proteases, extracellular LipA lipase, and hemolytic PlcH phospholipase C [52]. Fur is a global regulator that controls the expression of hundreds of genes involved not only in iron acquisition and virulence but also in some other aspects of P. aeruginosa biology such as respiration, metabolism, and stress responses [52].This wide spectrum of Fur targets explains why this protein is essential in P. aeruginosa for growth on solid media, especially considering the central role it plays in iron homeostasis [17][50].
5.2. PchR
PchR belongs to the AraC family of transcriptional regulators and activates the transcription of gene clusters pchDCBA and pchEF for pyochelin synthesis and fptABCX for pyochelin uptake [17]. The iron-loaded form of pyochelin acts as an effector molecule, activating the gene transcription of PchR targets, while PchR represses its own expression when siderophore is absent [17]. Pyochelin was shown to enhance growth and lethality of P. aeruginosa during infections, and a mutant defective in pyochelin and pyoverdine production was shown to be less virulent than a single-pyoverdine mutant [53]. In addition to pyochelin production, PchR controls the expression of the pqsABCDE gene cluster for PQS production and a number of virulence factors related to QS, such as the elastase lasB gene, the lectin lecA and lecB genes, and the gene clusters phzA1-G1 and hcnABC for phenazine and hydrogen cyanide production, respectively [54]. PchR is also involved in regulating the expression of other virulence-related genes, such as the algD gene for alginate production, the fimV gene for the synthesis of type IVa pili, and the bacterioferritin brfB gene [54].
5.3. AmpR
The β-lactam resistance regulator AmpR belongs to the LysR family of transcriptional regulators, and it is an important regulator of iron acquisition and the oxidative stress response [55]. AmpR acts as a global regulator, controlling the expression of more than 500 genes [55]. AmpR promotes iron acquisition by activating the transcription of regulatory anti-sense sRNA asPrrF1 (antagonizing the sRNA PrrF1), as well as the pyoverdine and pyochelin biosynthesis genes and the hemophore hasAp gene [55]. The oxidative stress response is mediated via AmpR by the activation of the katA catalase gene and the hydrogen peroxide resistance rgRgsA sRNA [55]. Consequently, an ampR mutation results in impaired growth under iron-limiting conditions and under H2O2-induced oxidative stress [55]. Moreover, AmpR is critical for P. aeruginosa to survive treatment with β-lactam antibiotics, and it also contributes to increasing P. aeruginosa’s “evolvability”, raising the chances of strengthening or developing new antibiotic resistances [56]. Under physiological conditions, AmpR is repressed by the cell wall precursor UDP-MurNAc pentapeptide. When β-lactam antibiotics cause cell wall damage, the muropeptides resulting from its restoration are internalized and bind AmpR to activate the transcription of the ampC β-lactamase gene [57]. Upon activation, AmpR promotes the expression of other key virulent determinants such as the HHQ transporter mexEF-oprN genes, the phz and hcn gene clusters for the production of pyocyanin and hydrogen cyanide, respectively, and several genes from T3SS and T6SS [55][58]. Accordingly, a P. aeruginosa ampR mutant was found to reduce mortality in the C. elegans infection model [58].
References
- Ulrich, L.E.; Koonin, E.V.; Zhulin, I.B. One-Component Systems Dominate Signal Transduction in Prokaryotes. Trends Microbiol. 2005, 13, 52–56.
- Cashin, P.; Goldsack, L.; Hall, D.; O’Toole, R. Contrasting Signal Transduction Mechanisms in Bacterial and Eukaryotic Gene Transcription. FEMS Microbiol. Lett. 2006, 261, 155–164.
- Gumerov, V.M.; Ortega, D.R.; Adebali, O.; Ulrich, L.E.; Zhulin, I.B. MiST 3.0: An Updated Microbial Signal Transduction Database with an Emphasis on Chemosensory Systems. Nucleic Acids Res. 2020, 48, D459.
- Lee, J.; Zhang, L. The Hierarchy Quorum Sensing Network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41.
- Gilbert, K.B.; Kim, T.H.; Gupta, R.; Greenberg, E.P.; Schuster, M. Global Position Analysis of the Pseudomonas aeruginosa Quorum-Sensing Transcription Factor LasR. Mol. Microbiol. 2009, 73, 1072–1085.
- Rumbaugh, K.P.; Griswold, J.A.; Iglewski, B.H.; Hamood, A.N. Contribution of Quorum Sensing to the Virulence of Pseudomonas aeruginosa in Burn Wound Infections. Infect. Immun. 1999, 67, 5854–5862.
- Kang, H.; Gan, J.; Zhao, J.; Kong, W.; Zhang, J.; Zhu, M.; Li, F.; Song, Y.; Qin, J.; Liang, H. Crystal Structure of Pseudomonas aeruginosa RsaL Bound to Promoter DNA Reaffirms Its Role as a Global Regulator Involved in Quorum-Sensing. Nucleic Acids Res. 2017, 45, 699–710.
- Miranda, S.W.; Asfahl, K.L.; Dandekar, A.A.; Greenberg, E.P. Pseudomonas aeruginosa Quorum Sensing. Adv. Exp. Med. Biol. 2022, 1386, 95–115.
- Rumbaugh, K.P.; Griswold, J.A.; Hamood, A.N. Contribution of the Regulatory Gene LasR to the Pathogenesis of Pseudomonas aeruginosa Infection of Burned Mice. J. Burn Care Rehabil. 1999, 20, 42–49.
- Lesprit, P.; Faurisson, F.; Join-Lambert, O.; Roudot-Thoraval, F.; Foglino, M.; Vissuzaine, C.; Carbon, C. Role of the Quorum-Sensing System in Experimental Pneumonia Due to Pseudomonas aeruginosa in Rats. Am. J. Respir. Crit. Care Med. 2003, 167, 1478–1482.
- Ochsner, U.A.; Reiser, J. Autoinducer-Mediated Regulation of Rhamnolipid Biosurfactant Synthesis in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 1995, 92, 6424–6428.
- Mukherjee, S.; Moustafa, D.; Smith, C.D.; Goldberg, J.B.; Bassler, B.L. The RhlR Quorum-Sensing Receptor Controls Pseudomonas aeruginosa Pathogenesis and Biofilm Development Independently of Its Canonical Homoserine Lactone Autoinducer. PLoS Pathog. 2017, 13, e1006504.
- Pearson, J.P.; Feldman, M.; Iglewski, B.H.; Prince, A. Pseudomonas aeruginosa Cell-to-Cell Signaling Is Required for Virulence in a Model of Acute Pulmonary Infection. Infect. Immun. 2000, 68, 4331–4334.
- O’Loughlin, C.T.; Miller, L.C.; Siryaporn, A.; Drescher, K.; Semmelhack, M.F.; Bassler, B.L. A Quorum-Sensing Inhibitor Blocks Pseudomonas aeruginosa Virulence and Biofilm Formation. Proc. Natl. Acad. Sci. USA 2013, 110, 17981–17986.
- Déziel, E.; Gopalan, S.; Tampakaki, A.P.; Lépine, F.; Padfield, K.E.; Saucier, M.; Xiao, G.; Rahme, L.G. The Contribution of MvfR to Pseudomonas aeruginosa Pathogenesis and Quorum Sensing Circuitry Regulation: Multiple Quorum Sensing-Regulated Genes are Modulated without Affecting LasRI, RhlRI or the Production of N-Acyl-L-Homoserine Lactones. Mol. Microbiol. 2005, 55, 998–1014.
- Diggle, S.P.; Matthijs, S.; Wright, V.J.; Fletcher, M.P.; Chhabra, S.R.; Lamont, I.L.; Kong, X.; Hider, R.C.; Cornelis, P.; Cámara, M.; et al. The Pseudomonas aeruginosa 4-Quinolone Signal Molecules HHQ and PQS Play Multifunctional Roles in Quorum Sensing and Iron Entrapment. Chem. Biol. 2007, 14, 87–96.
- Sánchez-Jiménez, A.; Marcos-Torres, F.J.; Llamas, M.A. Mechanisms of Iron Homeostasis in Pseudomonas aeruginosa and Emerging Therapeutics Directed to Disrupt This Vital Process. Microb. Biotechnol. 2023, 16, 1475–1491.
- Zhao, J.; Yu, X.; Zhu, M.; Kang, H.; Ma, J.; Wu, M.; Gan, J.; Deng, X.; Liang, H. Structural and Molecular Mechanism of CdpR Involved in Quorum-Sensing and Bacterial Virulence in Pseudomonas aeruginosa. PLoS Biol. 2016, 14, e1002449.
- Baynham, P.J.; Ramsey, D.M.; Gvozdyev, B.V.; Cordonnier, E.M.; Wozniak, D.J. The Pseudomonas aeruginosa Ribbon-Helix-Helix DNA-Binding Protein AlgZ (AmrZ) Controls Twitching Motility and Biogenesis of Type IV Pili. J. Bacteriol. 2006, 188, 132–140.
- Baynham, P.J.; Wozniak, D.J. Identification and Characterization of AlgZ, an AlgT-Dependent DNA-Binding Protein Required for Pseudomonas aeruginosa AlgD Transcription. Mol. Microbiol. 1996, 22, 97–108.
- Jones, C.J.; Newsom, D.; Kelly, B.; Irie, Y.; Jennings, L.K.; Xu, B.; Limoli, D.H.; Harrison, J.J.; Parsek, M.R.; White, P.; et al. ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic Di-GMP Synthesis and Biofilm Development by Pseudomonas aeruginosa. PLoS Pathog. 2014, 10, e1003984.
- Kalia, M.; Resch, M.D.; Cherny, K.E.; Sauer, K. The Alginate and Motility Regulator AmrZ Is Essential for the Regulation of the Dispersion Response by Pseudomonas aeruginosa Biofilms. mSphere 2022, 7, 1–20.
- Tart, A.H.; Blanks, M.J.; Wozniak, D.J. The AlgT-Dependent Transcriptional Regulator AmrZ (AlgZ) Inhibits Flagellum Biosynthesis in Mucoid, Nonmotile Pseudomonas aeruginosa Cystic Fibrosis Isolates. J. Bacteriol. 2006, 188, 6483–6489.
- Gallegos, M.T.; Schleif, R.; Bairoch, A.; Hofmann, K.; Ramos, J.L. Arac/XylS Family of Transcriptional Regulators. Microbiol. Mol. Biol. Rev. 1997, 61, 393–410.
- Urbanowski, M.L.; Lykken, G.L.; Yahr, T.L. A Secreted Regulatory Protein Couples Transcription to the Secretory Activity of the Pseudomonas aeruginosa Type III Secretion System. Proc. Natl. Acad. Sci. USA 2005, 102, 9930–9935.
- Tian, Z.; Cheng, S.; Xia, B.; Jin, Y.; Bai, F.; Cheng, Z.; Jin, S.; Liu, X.; Wu, W. Pseudomonas aeruginosa ExsA Regulates a Metalloprotease, ImpA, That Inhibits Phagocytosis of Macrophages. Infect. Immun. 2019, 87, e69519.
- Urbanowski, M.L.; Brutinel, E.D.; Yahr, T.L. Translocation of ExsE into Chinese Hamster Ovary Cells Is Required for Transcriptional Induction of the Pseudomonas aeruginosa Type III Secretion System. Infect. Immun. 2007, 75, 4432–4439.
- McCaw, M.L.; Lykken, G.L.; Singh, P.K.; Yahr, T.L. ExsD Is a Negative Regulator of the Pseudomonas aeruginosa Type III Secretion Regulon. Mol. Microbiol. 2002, 46, 1123–1133.
- Karna, S.L.R.; Nguyen, J.Q.; Evani, S.J.; Qian, L.W.; Chen, P.; Abercrombie, J.J.; Sebastian, E.A.; Fourcaudot, A.B.; Leung, K.P. T3SS and Alginate Biosynthesis of Pseudomonas aeruginosa Impair Healing of Infected Rabbit Wounds. Microb. Pathog. 2020, 147, 104254.
- Allsopp, L.P.; Collins, A.C.Z.; Hawkins, E.; Wood, T.E.; Filloux, A. RpoN/Sfa2-Dependent Activation of the Pseudomonas aeruginosa H2-T6SS and Its Cognate Arsenal of Antibacterial Toxins. Nucleic Acids Res. 2022, 50, 227–243.
- Sana, T.G.; Soscia, C.; Tonglet, C.M.; Garvis, S.; Bleves, S. Divergent Control of Two Type VI Secretion Systems by RpoN in Pseudomonas aeruginosa. PLoS ONE 2013, 8, e76030.
- Bremer, E. Liberate and Grab It, Ingest and Digest It: The GbdR Regulon of the Pathogen Pseudomonas aeruginosa. J. Bacteriol. 2014, 196, 3–6.
- Germán Sánchez, D.; Primo, E.D.; Damiani, M.T.; Lisa, A.T. Pseudomonas aeruginosa GbdR Gene Is Transcribed from a Σ54-Dependent Promoter under the Control of NtrC/CbrB, IHF and BetI. Microbiology 2017, 163, 1343–1354.
- Wargo, M.J. Choline Catabolism to Glycine Betaine Contributes to Pseudomonas aeruginosa Survival during Murine Lung Infection. PLoS ONE 2013, 8, e56850.
- Pan, X.; Liang, H.; Zhao, X.; Zhang, Q.; Chen, L.; Yue, Z.; Yin, L.; Jin, Y.; Bai, F.; Cheng, Z.; et al. Regulatory and Structural Mechanisms of PvrA-Mediated Regulation of the PQS Quorum-Sensing System and PHA Biosynthesis in Pseudomonas aeruginosa. Nucleic Acids Res. 2023, 51, 2691–2708.
- Pan, X.; Fan, Z.; Chen, L.; Liu, C.; Bai, F.; Wei, Y.; Tian, Z.; Dong, Y.; Shi, J.; Chen, H.; et al. PvrA Is a Novel Regulator That Contributes to Pseudomonas aeruginosa Pathogenesis by Controlling Bacterial Utilization of Long Chain Fatty Acids. Nucleic Acids Res. 2021, 48, 5967–5985.
- Okino, N.; Ito, M. Molecular Mechanism for Sphingosine-Induced Pseudomonas Ceramidase Expression through the Transcriptional Regulator SphR. Sci. Rep. 2016, 6, 38797.
- LaBauve, A.E.; Wargo, M.J. Detection of Host-Derived Sphingosine by Pseudomonas aeruginosa Is Important for Survival in the Murine Lung. PLoS Pathog. 2014, 10, e1003889.
- Maddocks, S.E.; Oyston, P.C.F. Structure and Function of the LysR-Type Transcriptional Regulator (LTTR) Family Proteins. Microbiology 2008, 154, 3609–3623.
- Carty, N.L.; Layland, N.; Colmer-Hamood, J.A.; Calfee, M.W.; Pesci, E.C.; Hamood, A.N. PtxR Modulates the Expression of QS-Controlled Virulence Factors in the Pseudomonas aeruginosa Strain PAO1. Mol. Microbiol. 2006, 61, 782–794.
- Daddaoua, A.; Krell, T.; Ramos, J.L. Transcriptional Control by Two Interacting Regulatory Proteins: Identification of the PtxS Binding Site at PtxR. Nucleic Acids Res. 2013, 41, 10150–10156.
- Daddaoua, A.; Fillet, S.; Fernández, M.; Udaondo, Z.; Krell, T.; Ramos, J.L. Genes for Carbon Metabolism and the ToxA Virulence Factor in Pseudomonas aeruginosa are Regulated through Molecular Interactions of PtxR and PtxS. PLoS ONE 2012, 7, e39390.
- Hobman, J.L. MerR Family Transcription Activators: Similar Designs, Different Specificities. Mol. Microbiol. 2007, 63, 1275–1278.
- Dietrich, L.E.P.; Price-Whelan, A.; Petersen, A.; Whiteley, M.; Newman, D.K. The Phenazine Pyocyanin Is a Terminal Signalling Factor in the Quorum Sensing Network of Pseudomonas aeruginosa. Mol. Microbiol. 2006, 61, 1308–1321.
- Fujikawa, M.; Kobayashi, K.; Tsutsui, Y.; Tanaka, T.; Kozawa, T. Rational Tuning of Superoxide Sensitivity in SoxR, the Transcription Factor: Implications of Species-Specific Lysine Residues. Biochemistry 2017, 56, 403–410.
- Palma, M.; Zurita, J.; Ferreras, J.A.; Worgall, S.; Larone, D.H.; Shi, L.; Campagne, F.; Quadri, L.E.N. Pseudomonas aeruginosa SoxR Does Not Conform to the Archetypal Paradigm for SoxR-Dependent Regulation of the Bacterial Oxidative Stress Adaptive Response. Infect. Immun. 2005, 73, 2958–2966.
- Aendekerk, S.; Ghysels, B.; Cornelis, P.; Baysse, C. Characterization of a New Efflux Pump, MexGHI-OpmD, from Pseudomonas aeruginosa That Confers Resistance to Vanadium. Microbiology 2002, 148, 2371–2381.
- Sakhtah, H.; Koyama, L.; Zhang, Y.; Morales, D.K.; Fields, B.L.; Price-Whelan, A.; Hogan, D.A.; Shepard, K.; Dietrich, L.E.P. The Pseudomonas aeruginosa Efflux Pump MexGHI-OpmD Transports a Natural Phenazine That Controls Gene Expression and Biofilm Development. Proc. Natl. Acad. Sci. USA 2016, 113, E3538–E3547.
- Ha, U.; Jin, S. Expression of the SoxR Gene of Pseudomonas aeruginosa Is Inducible during Infection of Burn Wounds in Mice and Is Required To Cause Efficient Bacteremia. Infect. Immun. 1999, 67, 5324.
- Visca, P.; Imperi, F. An Essential Transcriptional Regulator: The Case of Pseudomonas aeruginosa Fur. Future Microbiol. 2018, 13, 853–856.
- Reinhart, A.A.; Nguyen, A.T.; Brewer, L.K.; Bevere, J.; Jones, J.W.; Kane, M.A.; Damron, F.H.; Barbier, M.; Oglesby-Sherrouse, A.G. The Pseudomonas aeruginosa PrrF Small RNAs Regulate Iron Homeostasis during Acute Murine Lung Infection. Infect. Immun. 2017, 85, e76416.
- Ochsner, U.A.; Wilderman, P.J.; Vasil, A.I.; Vasil, M.L. GeneChip Expression Analysis of the Iron Starvation Response in Pseudomonas aeruginosa: Identification of Novel Pyoverdine Biosynthesis Genes. Mol. Microbiol. 2002, 45, 1277–1287.
- Chadha, J.; Harjai, K.; Chhibber, S. Revisiting the Virulence Hallmarks of Pseudomonas aeruginosa: A Chronicle through the Perspective of Quorum Sensing. Environ. Microbiol. 2021, 24, 2630–2656.
- Huang, H.; Shao, X.; Xie, Y.; Wang, T.; Zhang, Y.; Wang, X.; Deng, X. An Integrated Genomic Regulatory Network of Virulence-Related Transcriptional Factors in Pseudomonas aeruginosa. Nat. Commun. 2019, 10, 2931.
- Balasubramanian, D.; Kumari, H.; Jaric, M.; Fernandez, M.; Turner, K.H.; Dove, S.L.; Narasimhan, G.; Lory, S.; Mathee, K. Deep Sequencing Analyses Expands the Pseudomonas aeruginosa AmpR Regulon to Include Small RNA-Mediated Regulation of Iron Acquisition, Heat Shock and Oxidative Stress Response. Nucleic Acids Res. 2014, 42, 979.
- Gifford, D.R.; Furió, V.; Papkou, A.; Vogwill, T.; Oliver, A.; MacLean, R.C. Identifying and Exploiting Genes That Potentiate the Evolution of Antibiotic Resistance. Nat. Ecol. Evol. 2018, 2, 1033–1039.
- Dik, D.A.; Domínguez-Gil, T.; Lee, M.; Hesek, D.; Byun, B.; Fishovitz, J.; Boggess, B.; Hellman, L.M.; Fisher, J.F.; Hermoso, J.A.; et al. Muropeptide Binding and the X-ray Structure of the Effector Domain of the Transcriptional Regulator AmpR of Pseudomonas aeruginosa. J. Am. Chem. Soc. 2017, 139, 1448.
- Balasubramanian, D.; Schneper, L.; Merighi, M.; Smith, R.; Narasimhan, G.; Lory, S.; Mathee, K. The Regulatory Repertoire of Pseudomonas aeruginosa AmpC SS-Lactamase Regulator AmpR Includes Virulence Genes. PLoS ONE 2012, 7, e34067.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
647
Revisions:
3 times
(View History)
Update Date:
05 Sep 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




