Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hyun-Min Kim | -- | 2761 | 2023-07-23 23:19:23 | | | |
| 2 | Lindsay Dong | -10 word(s) | 2751 | 2023-07-24 04:49:29 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kim, H.; Zheng, X.; Lee, E. Histone Modifiers and p53 in Regulating Gene Expression. Encyclopedia. Available online: https://encyclopedia.pub/entry/47150 (accessed on 08 February 2026).
Kim H, Zheng X, Lee E. Histone Modifiers and p53 in Regulating Gene Expression. Encyclopedia. Available at: https://encyclopedia.pub/entry/47150. Accessed February 08, 2026.
Kim, Hyun-Min, Xiaoyu Zheng, Ethan Lee. "Histone Modifiers and p53 in Regulating Gene Expression" Encyclopedia, https://encyclopedia.pub/entry/47150 (accessed February 08, 2026).
Kim, H., Zheng, X., & Lee, E. (2023, July 23). Histone Modifiers and p53 in Regulating Gene Expression. In Encyclopedia. https://encyclopedia.pub/entry/47150
Kim, Hyun-Min, et al. "Histone Modifiers and p53 in Regulating Gene Expression." Encyclopedia. Web. 23 July, 2023.
Copy Citation
Chromatin structure plays a fundamental role in regulating gene expression, with histone modifiers shaping the structure of chromatin by adding or removing chemical changes to histone proteins. The p53 transcription factor controls gene expression, binds target genes, and regulates their activity. While p53 has been extensively studied in cancer research, specifically in relation to fundamental cellular processes, including gene transcription, apoptosis, and cell cycle progression, its association with histone modifiers has received limited attention.
histone modifications
p53
cancer
gene regulation
chromatin structure
1. Introduction
Chromatin is the complex of DNA, histone proteins, and other associated proteins that make up the structure of chromosomes within the nucleus of eukaryotic cells. It is a well-organized and dynamic structure that plays a fundamental role in the packaging and regulating of DNA. The chromatin structure can either promote or inhibit gene expression, depending on the specific modifications present on the histones [1][2][3][4]. Enzymes known as histone modifiers are responsible for adding or removing chemical changes in the histone proteins, thereby shaping the chromatin structure. These modifications, such as acetylation, methylation, phosphorylation, and ubiquitination, exhibit diverse effects on chromatin structure and gene expression.
p53, as a transcription factor, plays a crucial role in controlling the expression of the various genes involved in cell cycle regulation, DNA repair, apoptosis, and other cellular processes [5][6][7]. It acts as a tumor suppressor by promoting cell cycle arrest or inducing apoptosis in response to DNA damage or cellular stress. Thus, p53 is one of the most crucial tumor suppressor genes in tumorigenesis. The activity of p53 is regulated by various mechanisms, including the post-translational modifications of both p53 and the histones surrounding it.
The interplay between histone modifiers, such as histone acetyltransferases/deacetylase and histone methyltransferases/demethylase, and p53 is crucial for regulating gene expression, maintaining genomic stability, facilitating chromatin remodeling, and promoting DNA repair. The synergy between the two ensures proper cellular responses to DNA damage, stress signals, and other regulatory cues [8][9][10][11][12][13][14][15][16]. p53 is the guardian of the genome, whose activity is implicated in most types of cancers, so its post-transcriptional modifications have been extensively studied [16][17][18]. However, the connection between histone modifiers and p53 gained less attention despite the growing body of research on the involvement of histone modifiers in cancer.
2. The Interplay between Histone Modifiers and p53
Multiple mechanisms contribute to the interplay between histone modifiers and p53. Firstly, histone modifiers indirectly influence p53 function by modifying the structure and accessibility of p53 target genes within the chromatin (Figure 1).
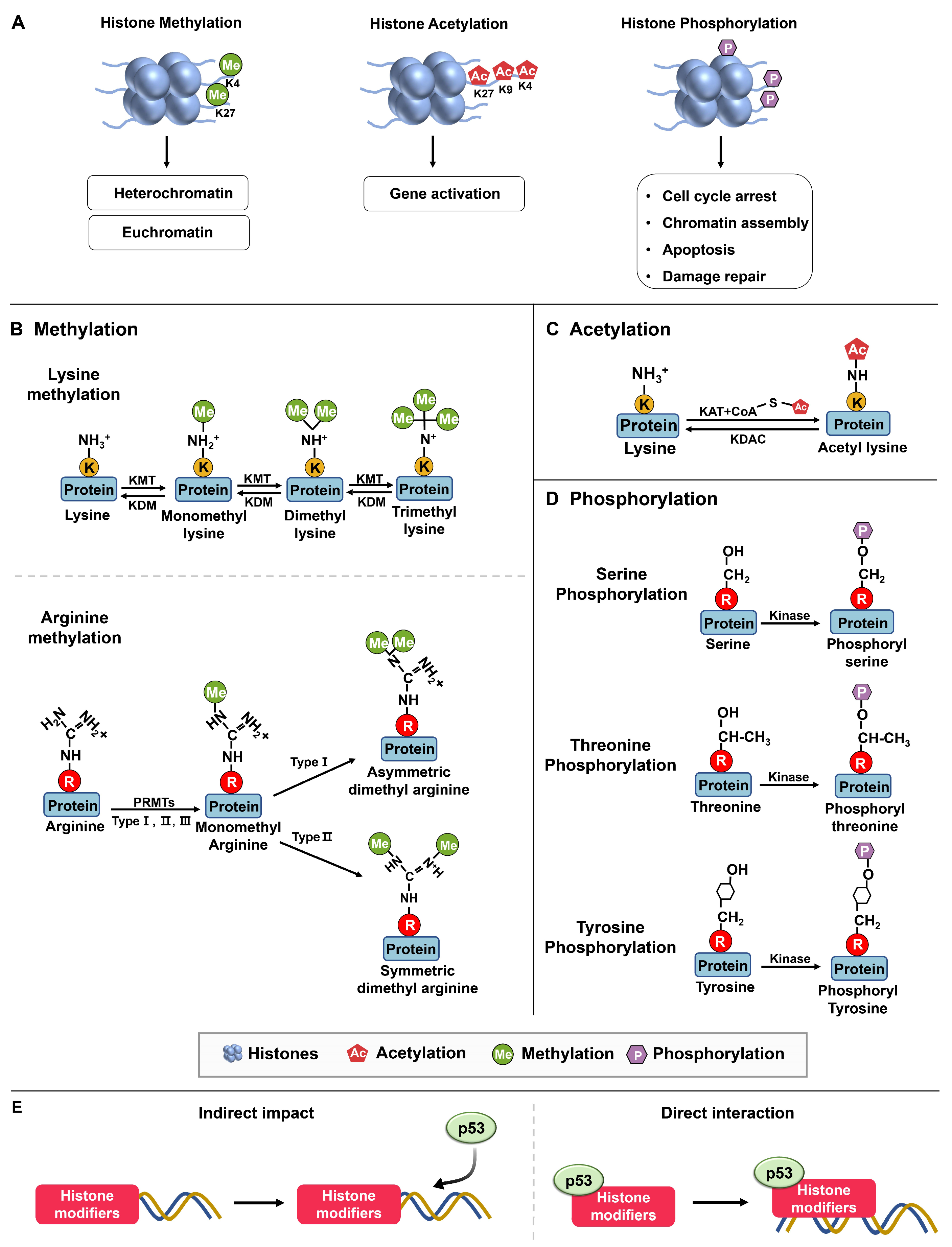
Figure 1. Histone modifications. (A) Methylation and demethylation are catalyzed by histone methyltransferase and histone demethylase, respectively. Euchromatin is characterized by specific molecular marks that indicate active gene expression, including histone acetylation, H3K4 trimethylation (H3K4me3), and H3K36 trimethylation (H3K36me3). In contrast, heterochromatin is marked by different modifications associated with gene repression and chromatin compaction. These marks include H3K9 trimethylation (H3K9me3) and H3K27 trimethylation (H3K27me3). Acetylation occurs in lysine residues catalyzed by histone acetyltransferase, while deacetylation is catalyzed by histone acetyltransferase. Phosphorylation: Kinases and phosphatases are enzymes involved in the addition and removal of phosphate groups, respectively, on proteins [19]. (B) Protein methylation occurs on lysine and arginine residues in histone and non-histone proteins through protein methyltransferases. The specific methyltransferases and demethylases reversibly regulated lysine methylation and demethylation from mono to trimethylation. Arginine methylations are induced in three types, including monomethylation, asymmetric dimethylation, and symmetric dimethylation [20]. (C) Acetyltransferases (KATs) transfer the acetyl group from acetyl–CoA to specific lysine residues in proteins, while acetylation can be reversed by lysine deacetylases (KDACs). (D) Protein phosphorylation occurs in serine, threonine, and tyrosine residues [21]. (E) Histone modifiers and p53 interact through mechanisms: Indirect chromatin modifications and direct recruitment target genes, affecting gene expression.
2.1. Histone Modifiers on p53
Packaging DNA into chromatin affects gene expression by making specific genes accessible to transcription factors, which are proteins that bind to DNA and control gene expression. Histone modifications affect chromatin structure by altering the interactions between histones and DNA. Each of these modifications can affect chromatin structure and gene expression differently. Modifications that are associated with active transcription, such as the acetylation of histone three and histone four (H3 and H4) or the di- or trimethylation (me) of H3K4, are commonly referred to as euchromatin modifications [22].
The enzymes responsible for regulating post-translational epigenetic modifications on histones have been categorized into four groups based on their roles: writers, erasers, readers, and movers [23]. Writers add changes to histones and include DNA methyltransferases (DNMTs), histone lysine methyltransferases (KMTs), and histone acetyltransferases (HATs). These modifications can affect the chromatin structure and gene expression by either promoting or repressing gene transcription. Therefore, they play a crucial role in establishing and maintaining the epigenetic marks on histones, which modulates gene expression.
Furthermore, histone modifications can be classified into major groups based on the type of modification and the amino acid residue being modified, including acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and ADP-ribosylation. Among these modifications, methylation, acetylation, and phosphorylation are the primary ones observed in the interplay between histones and p53 (Figure 1).
2.1.1. Methylation
The primary amino acids that are susceptible to methylation are arginine and lysine (Figure 1B). Arginine methylation involves the monomethylation, asymmetric dimethylation, or symmetric dimethylation of arginine residues, which are mediated by three types of protein arginine methyltransferases (PRMT type I, II, III), and this modification plays a role in modulating protein function and various cellular activities. On the other hand, lysine methylation leads to the formation of mono-, di-, or trimethylated lysine residues.
SET7/9
SET7/9, encoded by the SETD7 gene, was independently discovered in 2001 by Reinberg’s lab (named SET9) and Zhang’s lab (named SET7) [24][25][26]. Initially recognized as a methyltransferase involved in the methylation of H3K4, SET7/9 facilitates transcriptional activation by displacing the histone deacetylase NuRD complex (HDAC) [25]. Moreover, the observation that SET7/9-mediated H3K4 methylation enhances histone acetylation, which is associated with gene activation, implies that SET7/9 positively regulates transcription.
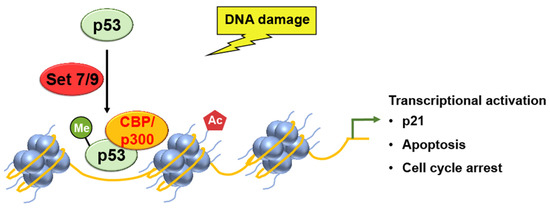
Ivanov et al. used chromatin immunoprecipitation (ChIP) assays to analyze the levels of p53 binding and histone H4 acetylation at the promoter of the p21 gene [27]. Methylation of p53, by SET7/9, increased the binding of p53 to the promoter region of the p21 gene, resulting in the increased acetylation of histone H4 and the subsequent transcriptional activation and cell cycle arrest (Figure 2).

Figure 2. SET7/9 methylation activates p53, which leads to the transcriptional activation of p21 gene expression, as well as p53-mediated apoptosis and cell cycle arrest.
G9a
G9a, a Set domain-containing protein, serves as the major histone lysine methyltransferase. It methylates histone H3 at both H3K9me1 and H3K9me2 modifications, earning its name G9a [28]. By adding methyl groups to histones, the G9a protein establishes and maintains epigenetic marks that regulate gene activity.
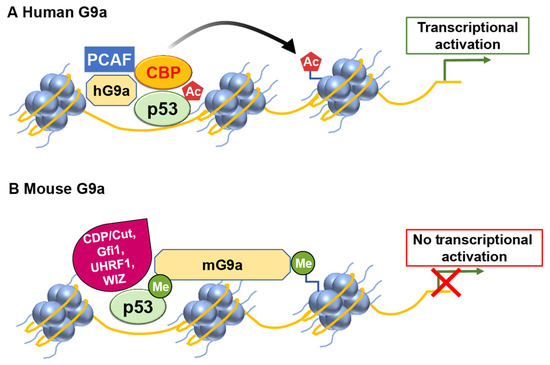
Human G9a (hG9a) can regulate the expression of p21 in a manner that is independent of p53 and its methylation activity [29]. G9a positively regulates p21 expression independently of p53 and its histone methyltransferase activity. Oh et al. demonstrated that hG9a upregulates p21 via interaction with PCAF, and this activating complex is recruited to the p21 promoter upon DNA damage-inducing agent etoposide treatment. Ultimately, p21 induction by G9a inhibits cellular proliferation and leads to apoptosis in p53-null cells. This regulatory mechanism does not rely on the histone–lysine methyltransferase activity of G9a and functions through a pathway separate from p53 (Figure 3).

Figure 3. G9a has the potential to modulate p53 transcriptional activity in a differential manner. (A) In humans, G9a functions as a coactivator for p53 by recruiting histone acetyltransferases (HATs) such as CBP and PCAF. (B) However, mouse G9a exerts a repressive effect on p53 transcriptional activity.
PRMTs (Protein Arginine Methyltransferases)
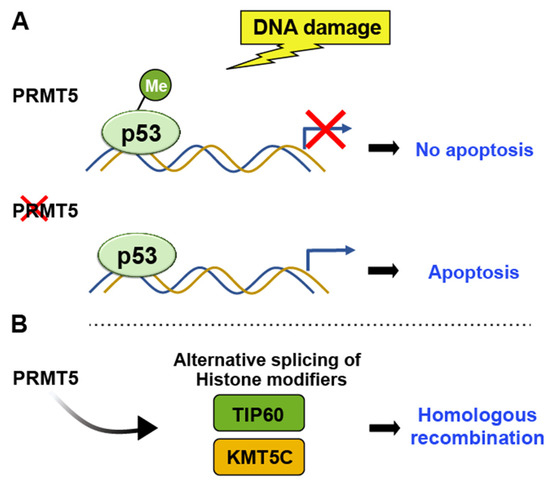
PRMTs have crucial roles in various cellular processes, including transcriptional regulation, chromatin regulation, signal transduction, and DNA damage repair. They catalyze the transfer of a methyl group from S-adenosylmethionine to the guanidine nitrogen of arginine residues in proteins [30][31]. PRMT5 specifically methylates histone H4 at arginine 3 (H4R3), indirectly influencing p53 activity by affecting the transcriptional regulation of p53 target genes. Following DNA damage, PRMT5 methylates p53 at arginine residues R333, R335, and R337 [32]. Bypassing p53 through arginine methylation leads to apoptosis evasion and facilitates tumor growth [15] (Figure 4).

Figure 4. PRMT5 methylates H4R3, indirectly influencing p53 activity by affecting the transcriptional regulation of p53 target genes. (A) Arginine methylation-induced bypassing of p53 leads to the evasion of apoptosis and facilitates tumor growth, whereas the depletion of PRMT5 induces p53-mediated apoptosis. (B) PRMT5 controls the alternative splicing of key histone-modifying enzymes such as TIP60 and KMT5C, thereby influencing chromatin structure and influencing DNA repair pathway.
JMJD2
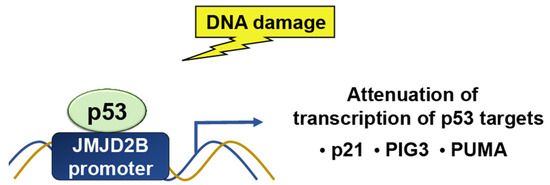
While p53 is a target for epigenetic modulators, it can also target histone modifiers. The Jumonji C domain, containing the histone demethylase 2 (JMJD2) family of proteins, selectively demethylates H3K9me3 and H3K36me3. JMJD2B/KDM4B is a p53-inducible gene in response to DNA damage (Figure 5). p53 regulates JMJD2B gene expression by binding to a p53-consensus motif in the JMJD2B promoter. JMJD2B induction attenuates the transcription of key p53 transcriptional targets, including p21, PIG3, and PUMA, while silencing enhances the induction of the two [33]. JMJD2B-mediated histone demethylation is also critical for p53-mediated autophagy and survival in Nutlin-treated cancer cells [14].

Figure 5. In response to DNA damage, p53 binds to a p53-consensus motif in the JMJD2B promoter; hence, p53 controls the expression of the JMJD2B gene. The induction of JMJD2B, in turn, suppresses the transcription of important p53 targets, such as p21, PIG3, and PUMA.
EZH2
EZH2, also known as Enhancer of Zeste Homolog 2, is a vital protein involved in epigenetic regulation. It belongs to the Polycomb group protein family and serves as the catalytic subunit of the Polycomb Repressive Complex 2 (PRC2) [34]. Functioning as a methyltransferase, EZH2 adds methyl groups, specifically, to lysine 27 of histone H3 (H3K27) through its histone methyltransferase activity. This enzymatic function enables EZH2 to modify chromatin structure by depositing the repressive histone mark H3K27me3.

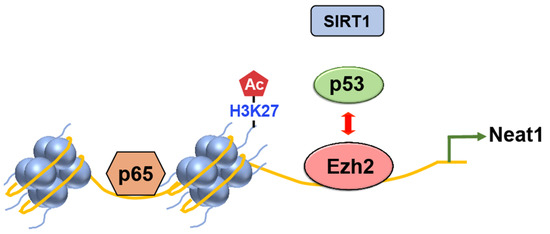
Figure 6. The competition between Ezh2 and p53 regulates inflammasome activation in mice. Upon exposure to inflammasome inducers, Ezh2 inhibits the binding of p53 to the promoter region of the lncRNA Neat1 gene. As a result, the recruitment of SIRT1 by p53 is also disrupted, preventing its binding to the DNA. This process leads to the enrichment of H3K27ac. Subsequently, the facilitated transcription of Neat1 by p65 promotes the activation of the inflammasome.
2.1.2. Phosphorylation
Phosphorylation is a common post-translational modification that involves adding a phosphate group (PO43−) to specific amino acid residues in proteins—typically serine, threonine, or tyrosine (Figure 1). This modification is catalyzed by protein kinases, which transfer the phosphate group from ATP to the target residue. Phosphorylation of p53 can occur at multiple sites in response to various stress signals. Phosphorylation of p53 at Ser15, in response to ionizing radiation, enhances the transcriptional activity of p53 by increasing its affinity for DNA to recruit coactivators such as CBP/p300 (Figure 7) [35][36].

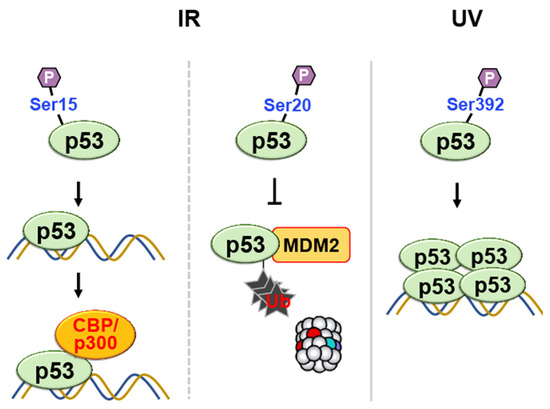
Figure 7. Phosphorylation of p53 at multiple sites in response to DNA damage regulates its transcriptional activity, DNA binding affinity, and protection against degradation. Phosphorylation at Ser15 and Ser20 enhances transcriptional activity and prevents ubiquitin-mediated degradation, respectively. Phosphorylation at Ser-392 enhances sequence-specific DNA binding and stabilizes tetramer formation following UV irradiation.
MAP Kinase Cascade
MAP kinase cascade is one of the major UV response pathways [37]. This pathway has three distinct components in mammalian cells: extracellular signal-regulated protein kinases (ERKs), p38 kinases, and stress-activated c-Jun N-terminal kinases (JNKs). These kinases participate in the regulation of cell proliferation, differentiation, stress responses, and apoptosis.
p38 MAP Kinase
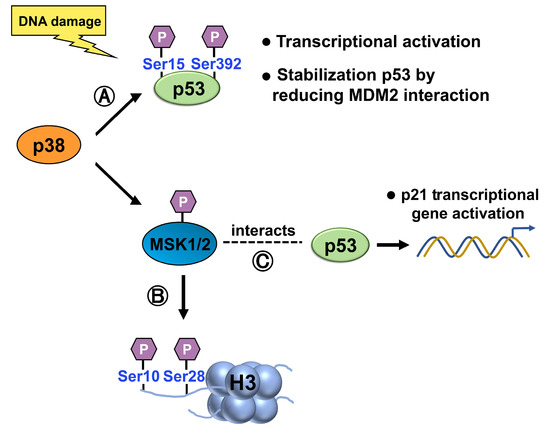
p38 can directly phosphorylate and activate p53. Upon activation, p38 phosphorylates specific serine residues on p53, such as Ser15 and Ser392 [37][38][39], leading to increased p53 stability, transcriptional activity, and the subsequent induction of downstream target genes involved in cell cycle arrest, DNA repair, and apoptosis (Figure 8). Ser15 phosphorylation also stabilizes p53 by reducing its interaction with MDM2, a negative regulatory partner [40]. Hence, phosphorylation of p53 is likely to play an essential role in regulating its activity.

Figure 8. (A) p38 phosphorylates p53, at Ser15 and Ser392, activating p53 and increasing its stability, transcriptional activity, and induction of target genes involved in cell cycle arrest, DNA repair, and apoptosis. Ser15 phosphorylation also stabilizes p53 by reducing its interaction with MDM2. (B) Upon activation by p38, 1/2 phosphorylates histone H3 at Ser10 and Ser28, modulating chromatin structure and gene expression. (C) When activated by the p38 MAPK pathway, MSK1 interacts with p53 and is recruited to the p21 promoter, where it phosphorylates histone H3 in a p53-dependent manner.
RSK2
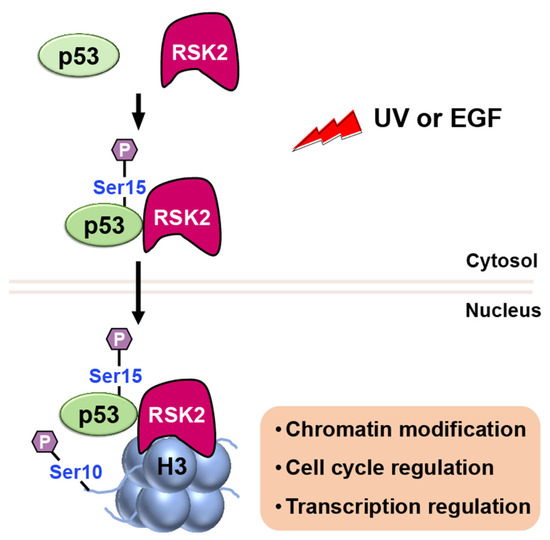
RSK2 is a p90 ribosomal S6 kinase family member that is activated by growth factors, peptide hormones, and neurotransmitters via MAPK/ERK signaling (ERK1 and ERK2). It is critical in regulating gene transcription by phosphorylating CBP at Ser133 [41]. Additionally, RSK2 has been reported to phosphorylate both histone H3 and p53 [42]. When cells are stimulated with UV or EGF, RSK2 is activated through the MAPK cascade, and it phosphorylates p53 protein at Ser15 (Figure 9) [42].

Figure 9. Upon stimulation with UV or EGF, activated RSK2 phosphorylates p53 at Ser15, and the RSK2/p53 complex translocates to the nucleus, where RSK2 phosphorylates histone H3 at Ser10, contributing to transcriptional regulation, chromatin remodeling, and cell cycle control.
2.1.3. Acetylation
Acetylation plays a significant role in modulating the transcriptional activity of p53, serving as a substantial modification (Figure 1) [43][44][45]. It is a post-translational modification that adds an acetyl group (-COCH3) to specific amino acid residues in proteins, predominantly lysine. This modification is catalyzed by enzymes known as histone acetyltransferases (HATs) or lysine acetyltransferases (KATs) [46].
CBP/p300
CREB-binding protein (CBP/p300), in mediating p53 acetylation and its consequential effect on p53 activity, has been illuminated in previous studies [47][48][49][50]. Extensive research has delved into unraveling the impact of acetylation on the regulation of p53’s functionality. The interaction between CBP/p300 and p53 leads to the acetylation of specific lysine residues within the regulatory part of p53, resulting in a conformational change that enhances its DNA binding activity [51].
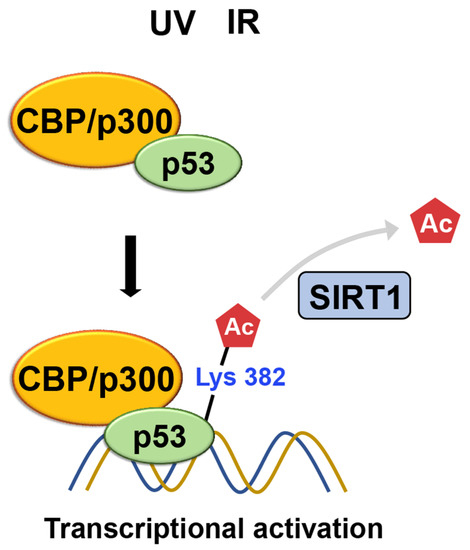
The ability of p53 to be acetylated was subsequently confirmed using acetylation-specific antibodies [52]. Sakaguchi et al. show that CBP/p300 acetylates K382 of p53 using a polyclonal antiserum, specific for p53, that is phosphorylated or acetylated at specific residues, while ATM phosphorylates S33 and S37 in response to UV irradiation. The acetylated p53 leads to increased binding to DNA. After DNA damage from irradiation, acetylation occurs at specific lysine residues, K382 and K320 of the p53, resulting in the recruitment of coactivators, such as CBP/p300 and TRRAP, to the p21 promoter and increasing histone acetylation. This suggests that a cascade of acetylation, in which p53-dependent recruitment of coactivators/HATs occurs, is essential for p53 to function correctly (Figure 10) [53].

Figure 10. Following DNA damage caused by irradiation, lysine residues of p53 undergo acetylation. This acetylation leads to the recruitment of coactivators such as CBP/p300 and TRRAP to the p21 promoter, thereby increasing histone acetylation. Meanwhile, SIRT1-mediated K382 deacetylation inhibits p53’s transcriptional activation, promoting its degradation.
SIRT1
SIRT1 was the first enzyme identified to target p53, which is not a histone, for deacetylation [54]. SIRT1 plays a role in regulating the cellular response to DNA damage by modifying the activity of p53 through the removal of acetyl groups from lysine residues [55][56][57][58][59]. Specifically, the deacetylation of K382 by SIRT1 inhibits the ability of p53 to activate transcription. This process leads to the degradation of p53, resulting in reduced apoptosis and increased cell survival when faced with DNA damage (Figure 10) [8][56].
TIP60
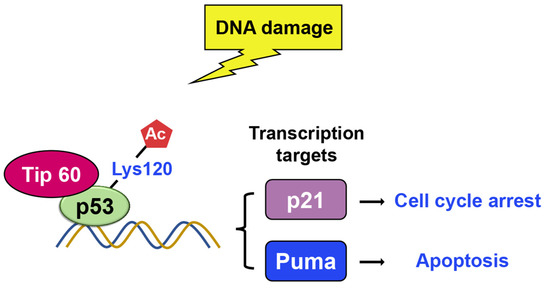
Tip60 is another histone acetylase linked to DNA damage repair and apoptosis [60][61][62][63][64][65][66]. There were two research groups who found that Tip60 induces K120 acetylation in the DNA binding domain upon DNA damage [61][67]. Lysine 120 (K120) acetylation occurs rapidly after DNA damage, and it is catalyzed by the MYST histone acetyltransferases hMOF and TIP60 (Figure 11) [67]. The mutation of K120 to arginine (K120R) debilitates K120 acetylation and blocks the transcription of pro-apoptotic target genes, such as BAX and PUMA, which, in turn, diminishes p53-mediated apoptosis without affecting cell cycle arrest. Additionally, the acetyl-K120 of p53 specifically accumulates at pro-apoptotic target genes. Additionally, studies indicate that Tip60–TRRAP complexes relocated to gamma-H2AX foci in response to DNA damage [63][68] and are crucial for the apoptotic response [64].

Figure 11. Upon DNA damage, Tip60 interacts with p53 and binds to its target gene promoters, leading to p21 activation and growth arrest. Additionally, Tip60 induces K120 acetylation, resulting in the activation of PUMA expression.
References
- Berger, S.L. Histone modifications in transcriptional regulation. Curr. Opin. Genet. Dev. 2002, 12, 142–148.
- Klemm, S.L.; Shipony, Z.; Greenleaf, W.J. Chromatin accessibility and the regulatory epigenome. Nat. Reviews. Genet. 2019, 20, 207–220.
- Sokolova, V.; Sarkar, S.; Tan, D. Histone variants and chromatin structure, update of advances. Comput. Struct. Biotechnol. J. 2023, 21, 299–311.
- Zaib, S.; Rana, N.; Khan, I. Histone Modifications and their Role in Epigenetics of Cancer. Curr. Med. Chem. 2022, 29, 2399–2411.
- Hafner, A.; Bulyk, M.L.; Jambhekar, A.; Lahav, G. The multiple mechanisms that regulate p53 activity and cell fate. Nat. Rev. Mol. Cell Biol. 2019, 20, 199–210.
- Hernandez Borrero, L.J.; El-Deiry, W.S. Tumor suppressor p53: Biology, signaling pathways, and therapeutic targeting. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188556.
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16.
- Yi, J.; Luo, J. SIRT1 and p53, effect on cancer, senescence and beyond. Biochim. Et Biophys. Acta 2010, 1804, 1684–1689.
- Huang, J.; Sengupta, R.; Espejo, A.B.; Lee, M.G.; Dorsey, J.A.; Richter, M.; Opravil, S.; Shiekhattar, R.; Bedford, M.T.; Jenuwein, T.; et al. p53 is regulated by the lysine demethylase LSD1. Nature 2007, 449, 105–108.
- Huang, J.; Perez-Burgos, L.; Placek, B.J.; Sengupta, R.; Richter, M.; Dorsey, J.A.; Kubicek, S.; Opravil, S.; Jenuwein, T.; Berger, S.L. Repression of p53 activity by Smyd2-mediated methylation. Nature 2006, 444, 629–632.
- Ren, X.; Tian, S.; Meng, Q.; Kim, H.M. Histone Demethylase AMX-1 Regulates Fertility in a p53/CEP-1 Dependent Manner. Front. Genet. 2022, 13, 929716.
- Kukita, A.; Sone, K.; Kaneko, S.; Kawakami, E.; Oki, S.; Kojima, M.; Wada, M.; Toyohara, Y.; Takahashi, Y.; Inoue, F.; et al. The Histone Methyltransferase SETD8 Regulates the Expression of Tumor Suppressor Genes via H4K20 Methylation and the p53 Signaling Pathway in Endometrial Cancer Cells. Cancers 2022, 14, 5367.
- Engeland, K. Cell cycle regulation: p53-p21-RB signaling. Cell Death Differ. 2022, 29, 946–960.
- Duan, L.; Perez, R.E.; Lai, X.; Chen, L.; Maki, C.G. The histone demethylase JMJD2B is critical for p53-mediated autophagy and survival in Nutlin-treated cancer cells. J. Biol. Chem. 2019, 294, 9186–9197.
- Li, Y.; Diehl, J.A. PRMT5-dependent p53 escape in tumorigenesis. Oncoscience 2015, 2, 700–702.
- DeHart, C.J.; Chahal, J.S.; Flint, S.J.; Perlman, D.H. Extensive post-translational modification of active and inactivated forms of endogenous p53. Mol. Cell Proteom. 2014, 13, 1–17.
- Dai, C.; Gu, W. p53 post-translational modification: Deregulated in tumorigenesis. Trends Mol. Med. 2010, 16, 528–536.
- Bode, A.M.; Dong, Z. Post-translational modification of p53 in tumorigenesis. Nat. Rev. Cancer 2004, 4, 793–805.
- Lee, H.T.; Oh, S.; Ro, D.H.; Yoo, H.; Kwon, Y.W. The Key Role of DNA Methylation and Histone Acetylation in Epigenetics of Atherosclerosis. J. Lipid Atheroscler. 2020, 9, 419–434.
- Blanc, R.S.; Richard, S. Arginine Methylation: The Coming of Age. Mol. Cell 2017, 65, 8–24.
- Keck, F.; Ataey, P.; Amaya, M.; Bailey, C.; Narayanan, A. Phosphorylation of Single Stranded RNA Virus Proteins and Potential for Novel Therapeutic Strategies. Viruses 2015, 7, 5257–5273.
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719.
- Bates, S.E. Epigenetic Therapies for Cancer. N. Engl. J. Med. 2020, 383, 650–663.
- Wang, H.; Cao, R.; Xia, L.; Erdjument-Bromage, H.; Borchers, C.; Tempst, P.; Zhang, Y. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol. Cell 2001, 8, 1207–1217.
- Nishioka, K.; Chuikov, S.; Sarma, K.; Erdjument-Bromage, H.; Allis, C.D.; Tempst, P.; Reinberg, D. Set9, a novel histone H3 methyltransferase that facilitates transcription by precluding histone tail modifications required for heterochromatin formation. Genes Dev. 2002, 16, 479–489.
- Daks, A.; Vasileva, E.; Fedorova, O.; Shuvalov, O.; Barlev, N.A. The Role of Lysine Methyltransferase SET7/9 in Proliferation and Cell Stress Response. Life 2022, 12, 362.
- Ivanov, G.S.; Ivanova, T.; Kurash, J.; Ivanov, A.; Chuikov, S.; Gizatullin, F.; Herrera-Medina, E.M.; Rauscher, F., 3rd; Reinberg, D.; Barlev, N.A. Methylation-acetylation interplay activates p53 in response to DNA damage. Mol. Cell Biol. 2007, 27, 6756–6769.
- Tachibana, M.; Sugimoto, K.; Fukushima, T.; Shinkai, Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J. Biol. Chem. 2001, 276, 25309–25317.
- Oh, S.T.; Kim, K.B.; Chae, Y.C.; Kang, J.Y.; Hahn, Y.; Seo, S.B. H3K9 histone methyltransferase G9a-mediated transcriptional activation of p21. FEBS Lett. 2014, 588, 685–691.
- Bedford, M.T.; Clarke, S.G. Protein arginine methylation in mammals: Who, what, and why. Mol. Cell 2009, 33, 1–13.
- Karkhanis, V.; Hu, Y.J.; Baiocchi, R.A.; Imbalzano, A.N.; Sif, S. Versatility of PRMT5-induced methylation in growth control and development. Trends Biochem. Sci. 2011, 36, 633–641.
- Jansson, M.; Durant, S.T.; Cho, E.C.; Sheahan, S.; Edelmann, M.; Kessler, B.; La Thangue, N.B. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008, 10, 1431–1439.
- Castellini, L.; Moon, E.J.; Razorenova, O.V.; Krieg, A.J.; von Eyben, R.; Giaccia, A.J. KDM4B/JMJD2B is a p53 target gene that modulates the amplitude of p53 response after DNA damage. Nucleic Acids Res. 2017, 45, 3674–3692.
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2006, 439, 871–874.
- Lambert, P.F.; Kashanchi, F.; Radonovich, M.F.; Shiekhattar, R.; Brady, J.N. Phosphorylation of p53 serine 15 increases interaction with CBP. J. Biol. Chem. 1998, 273, 33048–33053.
- Grossman, S.R. p300/CBP/p53 interaction and regulation of the p53 response. Eur. J. Biochem. 2001, 268, 2773–2778.
- Dhanasekaran, N.; Premkumar Reddy, E. Signaling by dual specificity kinases. Oncogene 1998, 17, 1447–1455.
- She, Q.B.; Chen, N.; Dong, Z. ERKs and p38 kinase phosphorylate p53 protein at serine 15 in response to UV radiation. J. Biol. Chem. 2000, 275, 20444–20449.
- Cox, M.L.; Meek, D.W. Phosphorylation of serine 392 in p53 is a common and integral event during p53 induction by diverse stimuli. Cell Signal 2010, 22, 564–571.
- Shieh, S.Y.; Ikeda, M.; Taya, Y.; Prives, C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell 1997, 91, 325–334.
- Xing, J.; Ginty, D.D.; Greenberg, M.E. Coupling of the RAS-MAPK pathway to gene activation by RSK2, a growth factor-regulated CREB kinase. Science 1996, 273, 959–963.
- Cho, Y.Y.; He, Z.; Zhang, Y.; Choi, H.S.; Zhu, F.; Choi, B.Y.; Kang, B.S.; Ma, W.Y.; Bode, A.M.; Dong, Z. The p53 protein is a novel substrate of ribosomal S6 kinase 2 and a critical intermediary for ribosomal S6 kinase 2 and histone H3 interaction. Cancer Res. 2005, 65, 3596–3603.
- Bannister, A.J.; Kouzarides, T. Regulation of p53 by Histone Acetylation and Acetyltransferases. Biochem. Soc. Trans. 2011, 39, 349–355.
- Brooks, C.L.; Gu, W. The impact of acetylation and deacetylation on the p53 pathway. Protein Cell 2011, 2, 456–462.
- Gong, F.; Chiu, L.Y.; Miller, K.M. Acetylation Reader Proteins: Linking Acetylation Signaling to Genome Maintenance and Cancer. PLoS Genet. 2016, 12, e1006272.
- Shahbazian, M.D.; Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem. 2007, 76, 75–100.
- Gu, W.; Roeder, R.G. Activation of p53 sequence-specific DNA binding by acetylation of the p53 C-terminal domain. Cell 1997, 90, 595–606.
- Tang, Z.; Chen, W.Y.; Shimada, M.; Nguyen, U.T.T.; Kim, J.; Sun, X.J.; Sengoku, T.; McGinty, R.K.; Fernandez, J.P.; Muir, T.W.; et al. SET1 and p300 Act Synergistically, through Coupled Histone Modifications, in Transcriptional Activation by p53. Cell 2023, 186, 2280.
- Dornan, D.; Shimizu, H.; Perkins, N.D.; Hupp, T.R. DNA-dependent acetylation of p53 by the transcription coactivator p300. J. Biol. Chem. 2003, 278, 13431–13441.
- Wang, Y.H.; Tsay, Y.G.; Tan, B.C.; Lo, W.Y.; Lee, S.C. Identification and characterization of a novel p300-mediated p53 acetylation site, lysine 305. J. Biol. Chem. 2003, 278, 25568–25576.
- Lill, N.L.; Grossman, S.R.; Ginsberg, D.; DeCaprio, J.; Livingston, D.M. Binding and modulation of p53 by p300/CBP coactivators. Nature 1997, 387, 823–827.
- Sakaguchi, K.; Herrera, J.E.; Saito, S.; Miki, T.; Bustin, M.; Vassilev, A.; Anderson, C.W.; Appella, E. DNA damage activates p53 through a phosphorylation-acetylation cascade. Genes Dev. 1998, 12, 2831–2841.
- Barlev, N.A.; Liu, L.; Chehab, N.H.; Mansfield, K.; Harris, K.G.; Halazonetis, T.D.; Berger, S.L. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol. Cell 2001, 8, 1243–1254.
- Luo, J.; Nikolaev, A.Y.; Imai, S.; Chen, D.; Su, F.; Shiloh, A.; Guarente, L.; Gu, W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell 2001, 107, 137–148.
- Sasca, D.; Hähnel, P.S.; Szybinski, J.; Khawaja, K.; Kriege, O.; Pante, S.V.; Bullinger, L.; Strand, S.; Strand, D.; Theobald, M.; et al. SIRT1 prevents genotoxic stress-induced p53 activation in acute myeloid leukemia. Blood 2014, 124, 121–133.
- Vaziri, H.; Dessain, S.K.; Ng Eaton, E.; Imai, S.I.; Frye, R.A.; Pandita, T.K.; Guarente, L.; Weinberg, R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 2001, 107, 149–159.
- Liu, T.; Lin, Y.H.; Leng, W.; Jung, S.Y.; Zhang, H.; Deng, M.; Evans, D.; Li, Y.; Luo, K.; Qin, B.; et al. A divergent role of the SIRT1-TopBP1 axis in regulating metabolic checkpoint and DNA damage checkpoint. Mol. Cell 2014, 56, 681–695.
- Chen, Y.; Zhang, H.; Xu, Z.; Tang, H.; Geng, A.; Cai, B.; Su, T.; Shi, J.; Jiang, C.; Tian, X.; et al. A PARP1-BRG1-SIRT1 axis promotes HR repair by reducing nucleosome density at DNA damage sites. Nucleic Acids Res. 2019, 47, 8563–8580.
- Alves-Fernandes, D.K.; Jasiulionis, M.G. The Role of SIRT1 on DNA Damage Response and Epigenetic Alterations in Cancer. Int. J. Mol. Sci. 2019, 20, 3153.
- Ikura, T.; Ogryzko, V.V.; Grigoriev, M.; Groisman, R.; Wang, J.; Horikoshi, M.; Scully, R.; Qin, J.; Nakatani, Y. Involvement of the TIP60 histone acetylase complex in DNA repair and apoptosis. Cell 2000, 102, 463–473.
- Tang, Y.; Luo, J.; Zhang, W.; Gu, W. Tip60-dependent acetylation of p53 modulates the decision between cell-cycle arrest and apoptosis. Mol. Cell 2006, 24, 827–839.
- Sun, Y.; Jiang, X.; Price, B.D. Tip60: Connecting chromatin to DNA damage signaling. Cell Cycle 2010, 9, 930–936.
- Murr, R.; Loizou, J.I.; Yang, Y.G.; Cuenin, C.; Li, H.; Wang, Z.Q.; Herceg, Z. Histone acetylation by Trrap-Tip60 modulates loading of repair proteins and repair of DNA double-strand breaks. Nat. Cell Biol. 2006, 8, 91–99.
- Tyteca, S.; Vandromme, M.; Legube, G.; Chevillard-Briet, M.; Trouche, D. Tip60 and p400 are both required for UV-induced apoptosis but play antagonistic roles in cell cycle progression. EMBO J. 2006, 25, 1680–1689.
- Wang, X.; Lupton, C.; Lauth, A.; Wan, T.C.; Foster, P.; Patterson, M.; Auchampach, J.A.; Lough, J.W. Evidence that the acetyltransferase Tip60 induces the DNA damage response and cell-cycle arrest in neonatal cardiomyocytes. J. Mol. Cell Cardiol. 2021, 155, 88–98.
- Gorrini, C.; Squatrito, M.; Luise, C.; Syed, N.; Perna, D.; Wark, L.; Martinato, F.; Sardella, D.; Verrecchia, A.; Bennett, S.; et al. Tip60 is a haplo-insufficient tumour suppressor required for an oncogene-induced DNA damage response. Nature 2007, 448, 1063–1067.
- Sykes, S.M.; Mellert, H.S.; Holbert, M.A.; Li, K.; Marmorstein, R.; Lane, W.S.; McMahon, S.B. Acetylation of the p53 DNA-binding domain regulates apoptosis induction. Mol. Cell 2006, 24, 841–851.
- Robert, F.; Hardy, S.; Nagy, Z.; Baldeyron, C.; Murr, R.; Dery, U.; Masson, J.Y.; Papadopoulo, D.; Herceg, Z.; Tora, L. The transcriptional histone acetyltransferase cofactor TRRAP associates with the MRN repair complex and plays a role in DNA double-strand break repair. Mol. Cell Biol. 2006, 26, 402–412.
More
Information
Subjects:
Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
782
Revisions:
2 times
(View History)
Update Date:
24 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




