
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Hera Hasan | -- | 4704 | 2023-07-19 09:03:53 | | | |
| 2 | Lindsay Dong | Meta information modification | 4704 | 2023-07-21 02:36:30 | | |
Video Upload Options
Glioblastoma is the most aggressive form of brain tumor originating from glial cells with a maximum life expectancy of 14.6 months. Despite the establishment of multiple promising therapies, the clinical outcome of glioblastoma patients is abysmal. Drug resistance has been identified as a major factor contributing to the failure of current multimodal therapy. Epigenetic modification, especially DNA methylation has been identified as a major regulatory mechanism behind glioblastoma progression. In addition, miRNAs, a class of non-coding RNA, have been found to play a role in the regulation as well as in the diagnosis of glioblastoma. The relationship between epigenetics, drug resistance, and glioblastoma progression has been clearly demonstrated. MGMT hypermethylation, leading to a lack of MGMT expression, is associated with a cytotoxic effect of TMZ in GBM, while resistance to TMZ frequently appears in MGMT non-methylated GBM.
1. The CNS5 Classification and Glioblastoma
2. DNA Methylation
2.1. DNA Methylation in GBM
2.2. DNA Methylation: Its Role in Regulation of Metabolism in GBM
2.3. PKM2
2.4. LDHA
2.5. HK2
3. miRNA
3.1. miRNA Biogenesis
3.2. miRNA in GBM
3.3. Upregulated miRNA
miR-21
3.4. miR-10b and miR-10a
3.5. miR-10b-miR-21
3.5.1. miR-10b and miR-222
3.5.2. miR-9
3.5.3. miR-221/222
3.5.4. miR-26a
3.5.5. miR-17-92 Cluster
3.5.6. miR-148a
3.5.7. Other Upregulated miRNA in GBM
3.6. Downregulated miRNAs
3.6.1. miR-31
3.6.2. miR-124
3.6.3. miR-34
3.6.4. miR-302-367 Cluster
3.6.5. miR-181
3.6.6. miR-219-5p and miR-219-1-3p
3.6.7. miR-1
3.6.8. miR-370-3p
3.6.9. miR-328
3.6.10. miR-375
3.6.11. miR-137
3.6.12. miR-128
3.6.13. miR-7
4. miRNA and DNA Methylation: An Epigenetic Interplay in GBM
5. miRNA and Epigenetic Modifications in TMZ Response and Drug Resistance
5.1. Epigenetic Modulation and TMZ Response
5.2. miRNA and TMZ Response
6. Diagnostic and Prognostic Molecular Tools in GBM
miRNAs are regulators of the pathways that play crucial roles in GBM invasion and progression. Their expression predicts the efficacy of conventional therapies that are routinely used in GBM treatment [26]. Most miRNAs have already been reported to be dysregulated in GBM so far. Therefore, miRNAs are currently being considered as potential diagnostic and prognostic biomarkers of gliomas [54]. Several studies have validated the potential roles of circulating miRNAs, particularly found in body fluids, such as CSF, plasma, and serum, in GBM diagnosis.
miR-21 is a potential biomarker of GBM with 90% sensitivity and 100% specificity [55]. It has been observed to have low expression in the post-operation serum of GBM patients, suggesting its potential as a serum-derived miRNA biomarker in GBM [33].
miR-26a and miR-21 are both circulatory miRNAs that are upregulated in GBM, and their serum expression levels have been observed to be reduced after surgery [33], suggesting their importance as candidate serum-based biomarkers in the diagnosis of GBM as well as in monitoring disease progression [33].
miR-10b is upregulated in GBM, and its overexpression promotes GBM progression and correlates with poor prognosis [55]. Its expression level positively correlates with WHO grades of gliomas as well as with tumor invasiveness [24]. Therefore, miR-10b might be used as a biomarker to evaluate glioma invasiveness and, subsequently, in the sub-classification of different tumor grades.
miR-328 is downregulated in GBM and acts as a tumor suppressor. The low expression level of miR-328 correlates with poor survival rate, thus it might be used as a candidate prognostic biomarker in GBM [45].
High plasma levels of miR-21 and low plasma levels of miR-128 and miR-342-3p act as candidate biomarkers in distinguishing GBM patients from healthy individuals with remarkably high sensitivity and specificity [56]. miR-342-3p expression is reduced in the plasma of glioma patients, and it is increased after surgery or chemotherapy.
miR-320a is a tumor suppressor miRNA, and its suppression correlates with excessive cell proliferation, invasion, and tumor growth [27]. Therefore, it might be used as a prognostic biomarker [27]. miR-146b and miR-4492 can be useful as novel biomarkers in predicting and monitoring GBM progression [27]. miR-146b is an oncogenic miRNA, and its major target is TRAF6. Downregulation of miR-146b and upregulation of TRAF6 correlate with inhibition of cell proliferation as well as apoptosis of tumor cells due to a decrease in Ki-67 expression.
miR-29 plasma level serves as a potential biomarker to indicate malignancy and glioma progression from grades I-II to grades III-IV [57]. miR-454-3p serum expression levels have been found markedly increased in GBM patients, and its upregulation correlates with poor prognosis.
Sometimes, single miRNA profiling is not sufficient enough to predict glioma outcomes. In such cases, profiles of several miRNAs are suggested. Seven miRNAs, including miR-15b, miR-23a, miR-133a, miR-150, miR-197, miR-497, and miR-548b-5p, are all downregulated in grades II-IV glioma patients, and the combined expression profiling of these miRNAs might be taken as a candidate biomarker in the prediction of GBM malignancy [57].
miR-181 is widely reported to be downregulated in GBM, especially in the early stages of this tumor [57]. Therefore, miR-181 might be used as a candidate biomarker for early prediction as well as in the identification of tumor grade. miR-181b and miR-181c act as predictive biomarkers of TMZ response in GBM [57] and may also help in choosing patients who are suitable for adjuvant therapy [26].
miR-221/222 is found to be significantly upregulated in plasma samples of glioma patients [26][58], and its overexpression contributes to poor prognosis and low survival rates [58].
7. GBM Therapy
7.1. miRNA Based Glioma Therapy
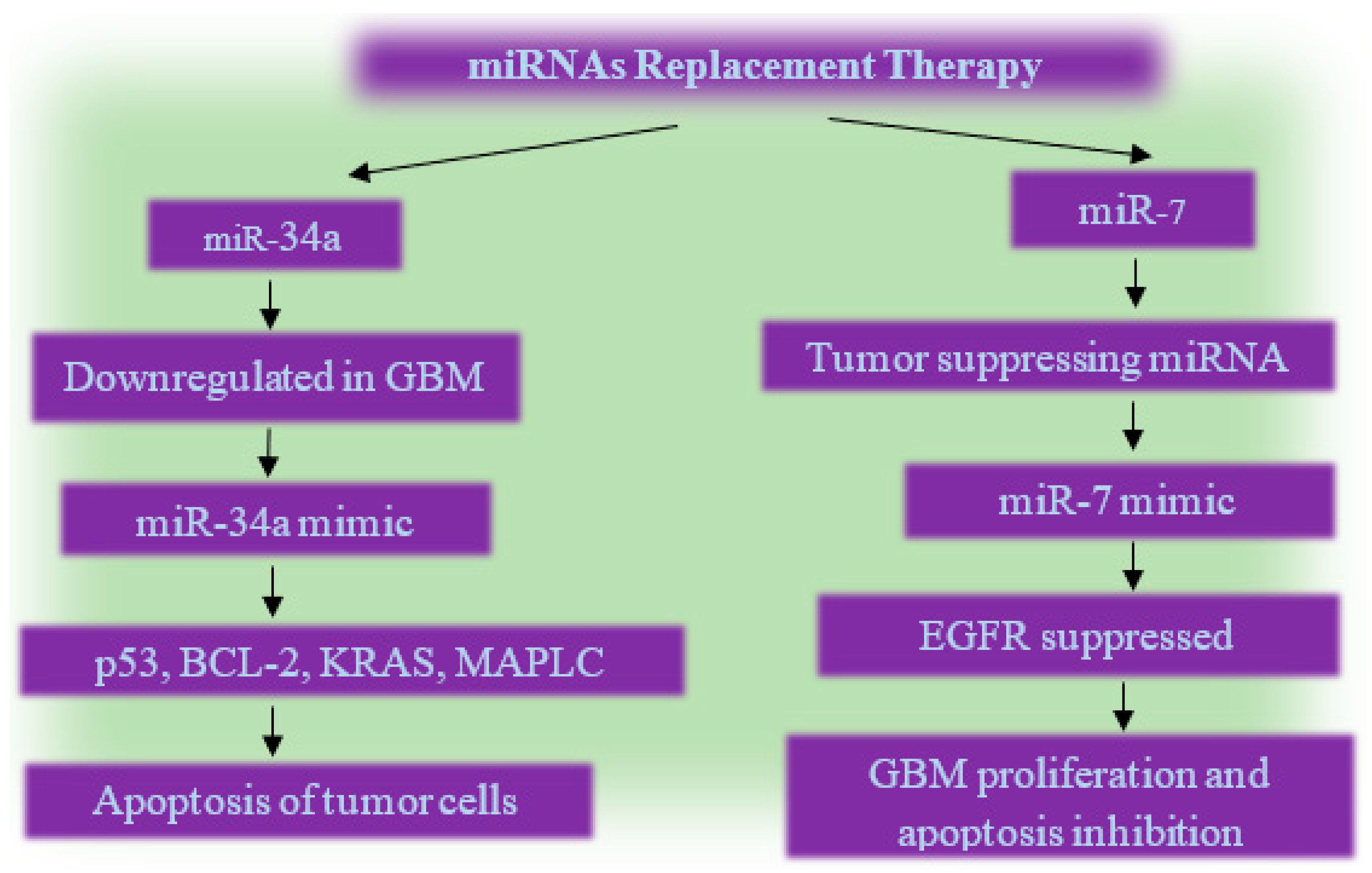
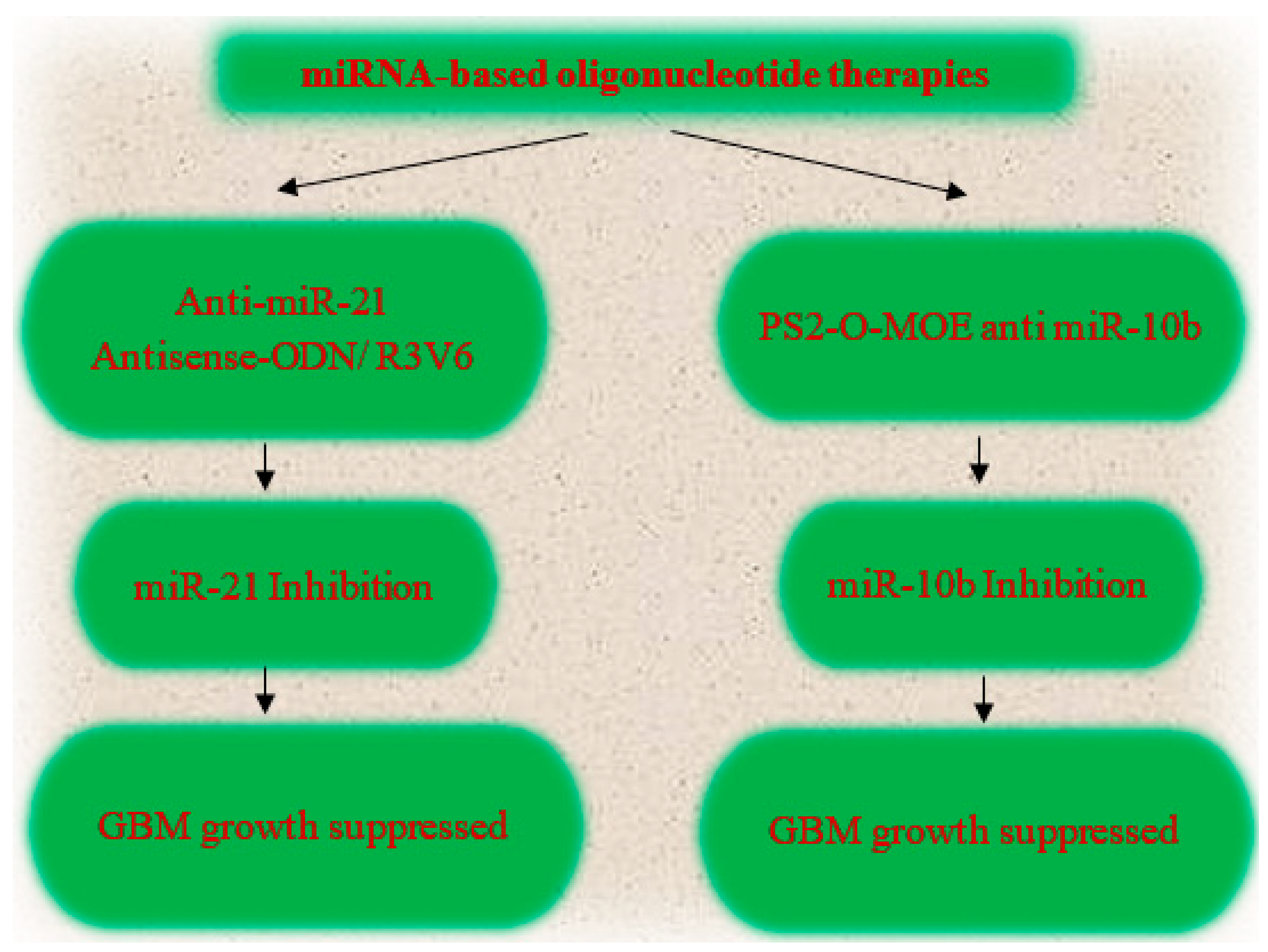
7.2. Epigenetic Therapy
7.3. Molecular Target Therapy
References
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251.
- Sejda, A.; Grajkowska, W.; Trubicka, J.; Szutowicz, E.; Wojdacz, T.; Kloc, W.; Iżycka-Świeszewska, E. WHO CNS5 2021 classification of gliomas: A practical review and road signs for diagnosing pathologists and proper patho-clinical and neuro-oncological cooperation. Folia Neuropathol. 2022, 60, 137–152.
- Wilson, T.A.; Karajannis, M.A.; Harter, D.H. Glioblastoma multiforme: State of the art and future therapeutics. Surg. Neurol. Int. 2014, 5, 64.
- Lombardi, M.Y.; Assem, M. Glioblastoma Genomics: A Very Complicated Story. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017.
- Mansouri, A.; Karamchandani, J.; Das, S. Molecular Genetics of Secondary Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Codon Publications: Brisbane, Australia, 2017.
- Burgess, R.; Jenkins, R.; Zhang, Z. Epigenetic changes in gliomas. Cancer Biol. 2008, 7, 1326–1334.
- Kreth, S.; Thon, N.; Kreth, F.W. Epigenetics in human gliomas. Cancer Lett. 2014, 342, 185–192.
- Ayala-Ortega, E.; Arzate-Mejia, R.; Perez-Molina, R.; Gonzalez-Buendia, E.; Meier, K.; Guerrero, G.; Recillas-Targa, F. Epigenetic silencing of miR-181c by DNA methylation in glioblastoma cell lines. BMC Cancer 2016, 16, 226.
- Marzese, D.M.; Hoon, D.S. Emerging technologies for studying DNA methylation for the molecular diagnosis of cancer. Expert Rev. Mol. Diagn. 2015, 15, 647–664.
- Quintavalle, C.; Mangani, D.; Roscigno, G.; Romano, G.; Diaz-Lagares, A.; Iaboni, M.; Donnarumma, E.; Fiore, D.; De Marinis, P.; Soini, Y.; et al. MiR-221/222 target the DNA methyltransferase MGMT in glioma cells. PLoS ONE 2013, 8, e74466.
- Dong, Z.; Cui, H. Epigenetic modulation of metabolism in glioblastoma. Semin. Cancer Biol. 2019, 57, 45–51.
- Chesnelong, C.; Chaumeil, M.M.; Blough, M.D.; Al-Najjar, M.; Stechishin, O.D.; Chan, J.A.; Pieper, R.O.; Ronen, S.M.; Weiss, S.; Luchman, H.A.; et al. Lactate dehydrogenase A silencing in IDH mutant gliomas. Neuro Oncol. 2014, 16, 686–695.
- Blakeway, D.; Karakoula, K.; Morris, M.; Rowther, F.; Eagles, L.; Darling, J.; Warr, T. Overexpression of Hexokinase 2 is epigenetically regulated by frequent hypomethylation in glioblastoma multiforme. Neuro Oncol. 2018, 20, i12.
- Kirstein, A.; Schmid, T.E.; Combs, S.E. The Role of miRNA for the Treatment of MGMT Unmethylated Glioblastoma Multiforme. Cancers 2020, 12, 1099.
- Rezaei, O.; Honarmand, K.; Nateghinia, S.; Taheri, M.; Ghafouri-Fard, S. miRNA signature in glioblastoma: Potential biomarkers and therapeutic targets. Exp. Mol. Pathol. 2020, 117, 104550.
- Henriksen, M.; Johnsen, K.B.; Andersen, H.H.; Pilgaard, L.; Duroux, M. MicroRNA expression signatures determine prognosis and survival in glioblastoma multiforme—A systematic overview. Mol. Neurobiol. 2014, 50, 896–913.
- Balandeh, E.; Mohammadshafie, K.; Mahmoudi, Y.; Hossein Pourhanifeh, M.; Rajabi, A.; Bahabadi, Z.R.; Mohammadi, A.H.; Rahimian, N.; Hamblin, M.R.; Mirzaei, H. Roles of Non-coding RNAs and Angiogenesis in Glioblastoma. Front. Cell Dev. Biol. 2021, 9, 716462.
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of MicroRNA Biogenesis, Mechanisms of Actions, and Circulation. Front. Endocrinol. 2018, 9, 402.
- Marumoto, T.; Saya, H. Molecular biology of glioma. Adv. Exp. Med. Biol. 2012, 746, 2–11.
- Michlewski, G.; Caceres, J.F. Post-transcriptional control of miRNA biogenesis. RNA 2019, 25, 1–16.
- Wu, K.; He, J.; Pu, W.; Peng, Y. The Role of Exportin-5 in MicroRNA Biogenesis and Cancer. Genom. Proteom. Bioinform. 2018, 16, 120–126.
- Uddin, M.S.; Mamun, A.A.; Alghamdi, B.S.; Tewari, D.; Jeandet, P.; Sarwar, M.S.; Ashraf, G.M. Epigenetics of glioblastoma multiforme: From molecular mechanisms to therapeutic approaches. Semin. Cancer Biol. 2022, 83, 100–120.
- Alfardus, H.; McIntyre, A.; Smith, S. MicroRNA Regulation of Glycolytic Metabolism in Glioblastoma. Biomed. Res. Int. 2017, 2017, 9157370.
- Kalkan, R.; Atli, E.I. The Impacts of miRNAs in Glioblastoma Progression. Crit. Rev. Eukaryot. Gene Expr. 2016, 26, 137–142.
- Hassan, A.; Mosley, J.; Singh, S.; Zinn, P.O. A Comprehensive Review of Genomics and Noncoding RNA in Gliomas. Top. Magn. Reson. Imaging 2017, 26, 3–14.
- Barciszewska, A.M. MicroRNAs as efficient biomarkers in high-grade gliomas. Folia Neuropathol. 2016, 54, 369–374.
- de Menezes, M.R.; Acioli, M.E.A.; da Trindade, A.C.L.; da Silva, S.P.; de Lima, R.E.; da Silva Teixeira, V.G.; Vasconcelos, L.R.S. Potential role of microRNAs as biomarkers in human glioblastoma: A mini systematic review from 2015 to 2020. Mol. Biol. Rep. 2021, 48, 4647–4658.
- Junior, L.G.D.; Baroni, M.; Lira, R.C.P.; Teixeira, S.; Fedatto, P.F.; Silveira, V.S.; Suazo, V.K.; Veronez, L.C.; Panepucci, R.A.; Antonio, D.S.M.; et al. High-throughput microRNA profile in adult and pediatric primary glioblastomas: The role of miR-10b-5p and miR-630 in the tumor aggressiveness. Mol. Biol. Rep. 2020, 47, 6949–6959.
- Sun, B.; Zhao, X.; Ming, J.; Liu, X.; Liu, D.; Jiang, C. Stepwise detection and evaluation reveal miR-10b and miR-222 as a remarkable prognostic pair for glioblastoma. Oncogene 2019, 38, 6142–6157.
- Coolen, M.; Katz, S.; Bally-Cuif, L. miR-9: A versatile regulator of neurogenesis. Front. Cell. Neurosci. 2013, 7, 220.
- Munoz, J.L.; Rodriguez-Cruz, V.; Rameshwar, P. High expression of miR-9 in CD133(+) glioblastoma cells in chemoresistance to temozolomide. J. Cancer Stem Cell Res. 2015, 3, e1003.
- Krichevsky, A.M.; Uhlmann, E.J. Oligonucleotide Therapeutics as a New Class of Drugs for Malignant Brain Tumors: Targeting mRNAs, Regulatory RNAs, Mutations, Combinations, and Beyond. Neurotherapeutics 2019, 16, 319–347.
- ParvizHamidi, M.; Haddad, G.; Ostadrahimi, S.; Ostadrahimi, N.; Sadeghi, S.; Fayaz, S.; Fard-Esfahani, P. Circulating miR-26a and miR-21 as biomarkers for glioblastoma multiform. Biotechnol. Appl. Biochem. 2019, 66, 261–265.
- Ghasemi, A.; Mohammadi, A.; Fallah, S. Epigenetic Modification of MicroRNA-219-1 and Its Association with Glioblastoma Multiforme. Biochemistry 2021, 86, 420–432.
- Ernst, A.; Campos, B.; Meier, J.; Devens, F.; Liesenberg, F.; Wolter, M.; Reifenberger, G.; Herold-Mende, C.; Lichter, P.; Radlwimmer, B. De-repression of CTGF via the miR-17-92 cluster upon differentiation of human glioblastoma spheroid cultures. Oncogene 2010, 29, 3411–3422.
- Cai, Q.; Zhu, A.; Gong, L. Exosomes of glioma cells deliver miR-148a to promote proliferation and metastasis of glioblastoma via targeting CADM1. Bull. Cancer 2018, 105, 643–651.
- Che, S.; Sun, T.; Wang, J.; Jiao, Y.; Wang, C.; Meng, Q.; Qi, W.; Yan, Z. miR-30 overexpression promotes glioma stem cells by regulating Jak/STAT3 signaling pathway. Tumour Biol. 2015, 36, 6805–6811.
- Lai, N.S.; Wu, D.G.; Fang, X.G.; Lin, Y.C.; Chen, S.S.; Li, Z.B.; Xu, S.S. Serum microRNA-210 as a potential noninvasive biomarker for the diagnosis and prognosis of glioma. Br. J. Cancer 2015, 112, 1241–1246.
- Liu, X.; Kang, J.; Sun, S.; Luo, Y.; Ji, X.; Zeng, X.; Zhao, S. iASPP, a microRNA-124 target, is aberrantly expressed in astrocytoma and regulates malignant glioma cell migration and viability. Mol. Med. Rep. 2018, 17, 1970–1978.
- Bazzoni, R.; Bentivegna, A. Role of Notch Signaling Pathway in Glioblastoma Pathogenesis. Cancers 2019, 11, 292.
- Li, Y.; Guessous, F.; Zhang, Y.; Dipierro, C.; Kefas, B.; Johnson, E.; Marcinkiewicz, L.; Jiang, J.; Yang, Y.; Schmittgen, T.D.; et al. MicroRNA-34a inhibits glioblastoma growth by targeting multiple oncogenes. Cancer Res. 2009, 69, 7569–7576.
- Janaki Ramaiah, M.; Divyapriya, K.; Kartik Kumar, S.; Rajesh, Y. Drug-induced modifications and modulations of microRNAs and long non-coding RNAs for future therapy against Glioblastoma Multiforme. Gene 2020, 723, 144126.
- Allen, B.K.; Stathias, V.; Maloof, M.E.; Vidovic, D.; Winterbottom, E.F.; Capobianco, A.J.; Clarke, J.; Schurer, S.; Robbins, D.J.; Ayad, N.G. Epigenetic pathways and glioblastoma treatment: Insights from signaling cascades. J. Cell. Biochem. 2015, 116, 351–363.
- Yang, C.H.; Wang, Y.; Sims, M.; Cai, C.; Pfeffer, L.M. MicroRNA-1 suppresses glioblastoma in preclinical models by targeting fibronectin. Cancer Lett. 2019, 465, 59–67.
- Wu, Z.; Sun, L.; Wang, H.; Yao, J.; Jiang, C.; Xu, W.; Yang, Z. MiR-328 expression is decreased in high-grade gliomas and is associated with worse survival in primary glioblastoma. PLoS ONE 2012, 7, e47270.
- Li, G.F.; Cheng, Y.Y.; Li, B.J.; Zhang, C.; Zhang, X.X.; Su, J.; Wang, C.; Chang, L.; Zhang, D.Z.; Tan, C.L.; et al. miR-375 inhibits the proliferation and invasion of glioblastoma by regulating Wnt5a. Neoplasma 2019, 66, 350–356.
- Cardoso, A.M.; Morais, C.M.; Pena, F.; Marante, T.; Cunha, P.P.; Jurado, A.S.; Pedroso de Lima, M.C. Differentiation of glioblastoma stem cells promoted by miR-128 or miR-302a overexpression enhances senescence-associated cytotoxicity of axitinib. Hum. Mol. Genet. 2021, 30, 160–171.
- Kelly, T.K.; De Carvalho, D.D.; Jones, P.A. Epigenetic modifications as therapeutic targets. Nat. Biotechnol. 2010, 28, 1069–1078.
- Banelli, B.; Forlani, A.; Allemanni, G.; Morabito, A.; Pistillo, M.P.; Romani, M. MicroRNA in glioblastoma: An overview. Int. J. Genom. 2017, 2017, 7639084.
- Morita, S.; Horii, T.; Kimura, M.; Ochiya, T.; Tajima, S.; Hatada, I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int. J. Mol. Sci. 2013, 14, 14647–14658.
- Bier, A.; Giladi, N.; Kronfeld, N.; Lee, H.K.; Cazacu, S.; Finniss, S.; Xiang, C.; Poisson, L.; deCarvalho, A.C.; Slavin, S.; et al. MicroRNA-137 is downregulated in glioblastoma and inhibits the stemness of glioma stem cells by targeting RTVP-1. Oncotarget 2013, 4, 665–676.
- Hiddingh, L.; Raktoe, R.S.; Jeuken, J.; Hulleman, E.; Noske, D.P.; Kaspers, G.J.; Vandertop, W.P.; Wesseling, P.; Wurdinger, T. Identification of temozolomide resistance factors in glioblastoma via integrative miRNA/mRNA regulatory network analysis. Sci. Rep. 2014, 4, 5260.
- Sun, J.; Ma, Q.; Li, B.; Wang, C.; Mo, L.; Zhang, X.; Tang, F.; Wang, Q.; Yan, X.; Yao, X.; et al. RPN2 is targeted by miR-181c and mediates glioma progression and temozolomide sensitivity via the wnt/β-catenin signaling pathway. Cell Death Dis. 2020, 11, 890.
- Wang, Q.; Li, P.; Li, A.; Jiang, W.; Wang, H.; Wang, J.; Xie, K. Plasma specific miRNAs as predictive biomarkers for diagnosis and prognosis of glioma. J. Exp. Clin. Cancer Res. 2012, 31, 97.
- Buruiana, A.; Florian, S.I.; Florian, A.I.; Timis, T.L.; Mihu, C.M.; Miclaus, M.; Osan, S.; Hrapsa, I.; Cataniciu, R.C.; Farcas, M.; et al. The Roles of miRNA in Glioblastoma Tumor Cell Communication: Diplomatic and Aggressive Negotiations. Int. J. Mol. Sci. 2020, 21, 1950.
- Zhang, Y.; Cruickshanks, N.; Pahuski, M.; Yuan, F.; Dutta, A.; Schiff, D.; Purow, B.; Abounader, R. Noncoding RNAs in Glioblastoma. In Glioblastoma; De Vleeschouwer, S., Ed.; Exon Publications: Brisbane, Australia, 2017.
- Yang, L.; Ma, Y.; Xin, Y.; Han, R.; Li, R.; Hao, X. Role of the microRNA 181 family in glioma development. Mol. Med. Rep. 2018, 17, 322–329.
- Zhang, R.; Pang, B.; Xin, T.; Guo, H.; Xing, Y.; Xu, S.; Feng, B.; Liu, B.; Pang, Q. Plasma miR-221/222 Family as Novel Descriptive and Prognostic Biomarkers for Glioma. Mol. Neurobiol. 2016, 53, 1452–1460.
- Romani, M.; Pistillo, M.P.; Banelli, B. Epigenetic Targeting of Glioblastoma. Front. Oncol. 2018, 8, 448.
- Mollaei, H.; Safaralizadeh, R.; Rostami, Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019, 234, 12369–12384.




