Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Athena Gogali | -- | 2537 | 2023-07-19 08:29:24 | | | |
| 2 | Alfred Zheng | Meta information modification | 2537 | 2023-07-20 04:24:38 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Exarchos, K.P.; Gkrepi, G.; Kostikas, K.; Gogali, A. Artificial Intelligence Applications in Interstitial Lung Diseases. Encyclopedia. Available online: https://encyclopedia.pub/entry/46956 (accessed on 07 February 2026).
Exarchos KP, Gkrepi G, Kostikas K, Gogali A. Artificial Intelligence Applications in Interstitial Lung Diseases. Encyclopedia. Available at: https://encyclopedia.pub/entry/46956. Accessed February 07, 2026.
Exarchos, Konstantinos P., Georgia Gkrepi, Konstantinos Kostikas, Athena Gogali. "Artificial Intelligence Applications in Interstitial Lung Diseases" Encyclopedia, https://encyclopedia.pub/entry/46956 (accessed February 07, 2026).
Exarchos, K.P., Gkrepi, G., Kostikas, K., & Gogali, A. (2023, July 19). Artificial Intelligence Applications in Interstitial Lung Diseases. In Encyclopedia. https://encyclopedia.pub/entry/46956
Exarchos, Konstantinos P., et al. "Artificial Intelligence Applications in Interstitial Lung Diseases." Encyclopedia. Web. 19 July, 2023.
Copy Citation
Interstitial lung diseases (ILDs) comprise a rather heterogeneous group of diseases varying in pathophysiology, presentation, epidemiology, diagnosis, treatment and prognosis. In the majority of ILDs, imaging modalities and especially high-resolution Computed Tomography (CT) scans have been the cornerstone in patient diagnostic approach and follow-up. The intricate nature of ILDs and the accompanying data have led to an increasing adoption of artificial intelligence (AI) techniques, primarily on imaging data but also in genetic data, spirometry and lung diffusion, among others.
interstitial lung diseases
artificial intelligence
1. Introduction
Interstitial lung diseases (ILDs), also known as diffuse parenchymal lung diseases, is an umbrella term encompassing more than 300 conditions. The term interstitial refers to the tissue surrounding the air sacs (alveoli) in the lung, bounded by the capillary endothelium and the alveolar epithelium. Most ILDs are often characterized by inflammation and/or fibrosis causing progressive decline in lung function and impaired gas exchange; consequently, patients complain of shortness of breath, cough, fatigue and weight loss. The most common causes of ILDs include exposure to environmental toxins, viral infections and autoimmune conditions, while many are idiopathic.
The diagnosis of ILDs is often challenging due to the diversity of symptoms and the fact that they can mimic symptoms of other respiratory disorders. A thorough medical evaluation, including pulmonary function tests, consecutive imaging studies, bronchoscopy and even biopsy, is often necessary to make a definitive diagnosis. Even after successful diagnosis, most of these exams are regularly collected and reviewed by the treating physician. As for epidemiology, ILDs range from ultra rare to relatively common. Overall, they collectively affect a considerable number of patients, posing a significant disease burden.
Artificial intelligence (AI) is a relatively old term which has been attracting significant attention during the last few years. AI refers to the technologies that enable machines to perform advanced human-like functions, such as learning, analyzing, seeing, etc., and gain experience as more data become available. AI is a broad field encompassing many different disciplines such as computer science, statistics, software engineering and many others. Machine learning (ML) is a subfield of AI where statistical models are trained to “learn” patterns from complex data in order to perform a specific task. Figure 1 shows a flowchart of the learning process.
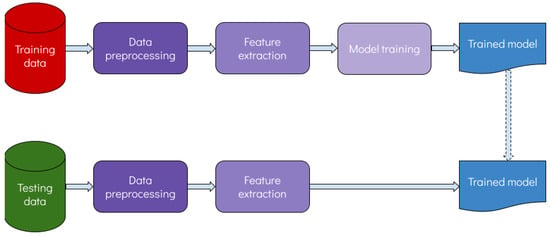
Figure 1. Flowchart of the learning process.
Advances in processing power and its wide availability in everyday devices such as home computers and smartphones have contributed significantly towards this direction. In addition, during the last few years, numerous machine learning and even deep learning software tools have been developed that require minimal or no coding skills to perform a certain task. All these advances go hand in hand with the production and storage of huge amounts of complex data from nearly every field of science and elsewhere. The most significant factor in the advent of ML and DL algorithms is the achievement of superior performance compared with traditional methods or algorithms. The ability to interpret or explain the output of some ML algorithms that have been traditionally considered “black-boxes” has certainly contributed towards wider acceptance of such algorithms.
This issue is specifically true for the utilization of ML algorithms in healthcare applications. Until recently, the lack of reasoning in medical decisions from ML algorithms has led to considerable skepticism that is being gradually alleviated. Moreover, it has become almost unanimously apparent that AI does not substitute medical personnel, but rather works in a decision-support manner, thus facilitating trivial tasks so that doctors can focus more freely and effectively on the patient. As a result, some impressive examples have been reported in the healthcare industry as well, especially with regard to computer vision tasks, at which DL algorithms are profoundly adept. Such examples include skin lesions [1], endoscopic images [2], histopathologic images [3] and radiology images. The latter type of data, especially CT scans, constitutes an integral part in the pursuit of ILDs. DL has revolutionized the way medical images are analyzed and interpreted and has been used in a wide range of medical imaging applications, including segmentation, registration, classification, object detection as well as many others. An exemplary architecture of a deep neural network is shown in Figure 2.
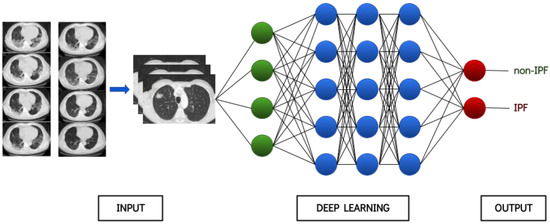
Figure 2. Provisional architecture of a deep neural network.
Besides imaging data, ILDs produce other sources of biomarkers that need to be analyzed in conjunction with the findings from the imaging modalities that are integral in the pursuit of ILDs. Recently, there is active interest in biomarkers coming from genomic, proteomic and transcriptomic investigations.
2. AI Applications in ILD Research
Applications of AI in ILD research can be roughly summarized in the following broad categories: (i) screening, (ii) diagnosis and classification, (iii) prognosis. The first category contains articles that aim to identify interstitial abnormalities before they become clinically meaningful and detectable. They focus primarily on interstitial lung abnormalities (ILAs) and exploit AI algorithms for their early identification, with the majority using CT scans as input. The second category, which is the most populated one, contains algorithms that diagnose ILDs mostly from imaging modalities, and some of them further aim to differentiate among various entities. The vast majority of studies in the last category (i.e., prognosis) deal with fibrosis and aim to assess its progression over time and subsequent prognosis. A summarizing table (Table 1) for the included studies has been added at the end of this section.
Table 1. Summary of the studies included in the current review article.
| Author(s) | Scope | Dataset | Type of Data | Performance |
|---|---|---|---|---|
| Bermejo-Peláez et al. [4] | Screening | 208 CT scans | Imaging | Sensitivity: 91.41% |
| Agarwala et al. [5] | Screening | 168 CT scans | Imaging | Success rate: 85.3% |
| Kim et al. [6] | Screening | 336 participants | Imaging | Accuracy: 90.5% |
| Nishikiori et al. [7] | Screening | 1159 chest X-rays | Imaging | AUC = 0.979 |
| Onishchenko et al. [8] | Screening | 2,983,215 participants | Electronic Health Records | AUC > 0.840 |
| Axelsson et al. [9] | Screening | >10,000 patients | Proteins | - |
| Pawar and Talbar [10] | Diagnosis & classification | 108 CT scans | Imaging | Accuracy: 89.39% |
| Huang et al. [11] | Diagnosis & classification | 108 CT scans | Imaging | F1-score > 0.96 |
| Chloe et al. [12] | Diagnosis & classification | 300 patients | Imaging | Accuracy: 60.9% |
| Koo et al. [13] | Diagnosis & classification | 1085 patients | Imaging | AUC > 0.900 |
| Furukawa et al. [14] | Diagnosis & classification | 1068 patients | Imaging | Accuracy: 83.6% |
| Christe et al. [15] | Diagnosis & classification | 105 patients | Imaging | Accuracy: 81% |
| Bratt et al. [16] | Diagnosis & classification | 1239 patients | Imaging | AUC = 0.870 |
| Yang et al. [17] | Diagnosis & classification | 1760 chest X-rays | Imaging | Accuracy: 92.46% |
| Horimasu et al. [18] | Diagnosis & classification | 60 patients | Auscultation | Accuracy: 75% |
| Plantier et al. [19] | Diagnosis & classification | 150 patients | Volatile Organic Compounds | Accuracy: 77.5% |
| Zhang et al. [20] | Diagnosis & classification | 300 patients | Gene Expression | - |
| Li et al. [21] | Diagnosis & classification | 600 patients | Gene Expression | AUC = 0.856 |
| Kim et al. [22] | Prognosis | 192 patients | Imaging | - |
| Handa et al. [23] | Prognosis | 465 patients | Imaging | - |
| Budzikowski et al. [24] | Prognosis | 169 patients | Imaging and Genomic | - |
| Liang et al. [25] | Prognosis | 116 patients | Imaging | AUC = 0.870 |
| Aoki et al. [26] | Prognosis | 104 patients | Imaging | - |
| Bowman et al. [27] | Prognosis | 589 patients | Proteomic | Sensitivity: 90% |
| Mayr et al. [28] | Prognosis | 124 patients | Proteomic | Accuracy: 83% |
2.1. Screening
The most common approach in this category involves the analysis of imaging modalities for the identification of ILAs. Bermejo-Peláez et al. [4] analyze 208 CT scans, broken down to 37,424 radiographic tissue samples, and utilize an ensemble of deep convolutional neural networks. Their aim is to identify radiographic patterns that precede the development of ILDs and classify them into eight different parenchymal feature classes, namely: normal parenchyma, five interstitial patterns (ground-glass, reticular, nodular, linear scar, subpleural line) and two emphysematous patterns (centrilobular and paraseptal). The authors report very good performance with an average sensitivity of 91.41% and average specificity of 98.18%. Agarwala et al. [5] employ 168 CT scans, coming both from a public database and a private one, and use a convolutional neural network (CNN) to identify three radiographic patterns (i.e., consolidation, emphysema and fibrosis), yielding acceptable performance, Similarly, Kim et al. [6] aim to identify ILAs in routine CT scans, exhibiting 90.5% overall accuracy. Since CT scans, and especially HRCTs (high-resolution CT scans) are not readily available in all facilities, Nishikiori et al. [7] propose a deep-learning-based algorithm which assigns scores to chest X-rays, representing the probability of fibrosing interstitial lung diseases. The performance of the algorithm was further validated with CT scans and its detection capability was not inferior to that of doctors (including pulmonologists and radiologists).
A methodologically different approach was proposed by Onishchenko et al. [8], who systematically searched electronic health records for comorbidity patterns that were potentially associated with the development of IPF. Interestingly, the proposed non-invasive methodology can predict IPF from 1 to 4 years prior to definite diagnosis, with Area under the ROC Curve (AUC) > 0.84. The proposed algorithm can be applied in large cohorts, even in primary care, enabling early diagnosis with potentially better outcomes.
Another interesting approach was presented by Axelsson et al. [9], who aimed to identify proteins in circulating blood that are associated with ILAs. More than 4700 protein analytes from two major databases with more than 10,000 patients combined were searched for potential association with the incidence and progression of ILAs. The extracted protein markers were subsequently coupled with machine learning algorithms in order to identify interstitial lung disease early and predict its progression.
2.2. Diagnosis and Classification
Some of the first articles to engage AI in ILD research comprise this category, which is also the most populated one. The majority utilize machine learning algorithms, and primarily deep-learning-based ones, in order to analyze CT scans and identify regions pertaining to interstitial lung diseases [10][11]. Pawar and Talbar [10] propose a two-stage approach: the first is for segmenting HRCT images and the second is for classifying segmented images into six ILD classes (normal, emphysema, fibrosis, ground glass, micronodules and consolidation). The proposed classifier yielded an overall accuracy of 89.39%. Huang et al. [11] employ a dataset consisting of 108 annotated HRCT images and classified regions in five ILD-relevant classes (healthy, ground glass, emphysema, micronodules and fibrosis) with an F1-score of more than 0.96.
Besides region classification, some studies go further to provide a specific diagnosis [12][13]. Chloe et al. [12] developed a content-based image retrieval algorithm in order to aid the diagnosis of ILDs. The algorithm was evaluated on images from approximately 300 patients belonging to four categories and yielded overall accuracy of 60.9%. The four categories were: usual interstitial pneumonia (UIP), nonspecific interstitial pneumonia (NSIP), cryptogenic organizing pneumonia, and chronic hypersensitivity pneumonitis. In a methodologically different approach but for the same purpose Koo et al. [13] achieved a slightly better performance.
Among ILDs, IPF attracts considerable interest since it is relatively frequent and also bears quite unfavorable prognosis. For this purpose various approaches have been presented in the literature [14][15][16]. In a similar manner with the previous approaches, these articles describe AI based approaches for segmenting and classifying images, with special focus on IPF related patterns. All three studies utilized deep learning algorithms and were developed with 1068, 105 and 1239 patients, reporting overall accuracy 83.6%, 81%, while the latter one [16] reported an AUC = 0.87. Yang et al. [17] focused on another popular ILD entity, i.e., pneumoconiosis, and analyzed 1760 chest X-rays with a deep learning algorithm in order to discriminate between patients with pneumoconiosis and normal ones. The best performing algorithm resulted in 92.46% accuracy, which is significantly higher compared to other machine learning algorithms (e.g., support vector machines, artificial neural networks, random forests, k-nearest neighbors) used in the same study.
A totally different approach was presented by Horimasu et al. [18], who developed a machine learning algorithm in order to discriminate fine crackles from other auscultatory sounds and subsequently associate them with respective regions of pulmonary fibrosis. In total, 60 patients underwent chest X-ray and HRCT, and respiratory findings were recorded in six positions. Fine crackles were mostly associated with honeycombing and traction bronchiectasis; another interesting finding was that the identification of fine crackles indicated a higher sensitivity compared to a chest-X-ray-based determination of the presence or absence of ILDs.
In a recently published pilot study, Plantier et al. [19] analyzed the exhaled air from three patient categories, namely, patients diagnosed with IPF, patients with ILD secondary to connective tissue diseases (CTDs) and healthy controls. The aim of the study was to identify volatile organic compound patterns in exhaled air to non-invasively discriminate IPF- and CTD-related ILDs. The study employed approximately 150 patients and resulted in classification accuracy of 77.5%, sensitivity of 76.5% and specificity of 78.4%.
Zhang et al. [20] utilized a machine learning algorithm aiming to identify biomarkers from gene expression profiles that differentiate chronic hypersensitivity pneumonitis (CHP) from other ILDs. Based on the analysis of approximately 300 patients, the authors reported 674 CHP biomarkers with an ultimate aim to facilitate precise gene therapy for CHP. In a similar approach that was recently published, Li et al. [21] identified a six-gene subset whose expression was significantly different in patients with IPF compared to healthy controls. A random forest algorithm was used upon a set of more than 600 patients, yielding an AUC of 0.856.
2.3. Prognosis
From another perspective, there is the use of AI and deep learning for the prognosis of the disease, either for the progression of ILD or the possibility of malignancy development or coexistence. Different tools were created mostly based on quantitative imaging from HRCT, and some quantitative interstitial scores have also been developed. Kim et al. [22] propose that the use of imaging features of quantitative lung fibrosis scores in patients with IPF in the early stages can predict the progressive fibrosis status later on. The ability to apply this in precisely informed and timely management decisions after treatment is progress. AI could be used in testing new safe and effective therapies and to elucidate the effects of therapies in patients with biologically heterogeneous disease in the timing of progression. New software such as the artificial-intelligence-based quantitative CT image analysis software (AIQCT) of Handa et al. quantified parenchymal lesions and airway volumes; they suggest that lung volume on HRCT imaging of the chest may provide additional prognostic information on the gender–age–lung physiology stage of IPF [23].
Since patients with idiopathic pulmonary fibrosis are at a higher risk of developing lung cancer, radiomics could be an early prognostic indicator. Budzikowski et al. correlate image features with patients’ genetic mutations [24]. It is stated that these imaging features may serve as prognostic indicators combining radiomic features and genetic mutations. That also provides an understanding of the interaction between imaging phenotype and patient genotype on the progression and, furthermore, the treatment of IPF. Additionally, regarding the risk of malignancy development or coexistence, in a recent study of Liang et al., it is mentioned that whole-lung CT texture analysis is a promising tool for the lung cancer risk stratification of IPF patients [25]. Moreover, Aoki et al. [26] using deep-learning-based analysis and measured consolidation with fibrosis, found that it was independently associated with poor survival. They also found that the lesion extent measured using deep-learning-based analysis showed a negative correlation with pulmonary function test results and prognosis.
Another domain that AI is applied to is the investigation of the proteomic signature of ILD. The exact correspondence of cell state changes in diseased organs to peripheral protein signatures currently remains unknown. Certain plasma biomarkers in patients with progressive fibrosing ILD have been identified, and consistent associations across ILD subtypes have been reported. In a recent research study [27], a proteomic signature comprising 12 biomarkers was derived via machine learning, suggesting that approximately 10% of patients with a low-risk proteomic signature would experience ILD progression in the year after blood draw. Finally, Mayr et al. [28], using cross-modal analysis and machine learning, identified the cellular source of biomarkers. They demonstrated that information transfer between modalities predicts disease status. So, they also suggest the feasibility of clinical cell state monitoring through the longitudinal sampling of body fluid proteomes.
References
- Gouda, W.; Sama, N.U.; Al-Waakid, G.; Humayun, M.; Jhanjhi, N.Z. Detection of Skin Cancer Based on Skin Lesion Images Using Deep Learning. Healthcare 2022, 10, 1183.
- Luo, X.; Zhang, J.; Li, Z.; Yang, R. Diagnosis of Ulcerative Colitis from Endoscopic Images Based on Deep Learning. Biomed. Signal Process. Control 2022, 73, 103443.
- Shamai, G.; Livne, A.; Polónia, A.; Sabo, E.; Cretu, A.; Bar-Sela, G.; Kimmel, R. Deep Learning-Based Image Analysis Predicts PD-L1 Status from H&E-Stained Histopathology Images in Breast Cancer. Nat. Commun. 2022, 13, 6753.
- Bermejo-Peláez, D.; Ash, S.Y.; Washko, G.R.; San José Estépar, R.; Ledesma-Carbayo, M.J. Classification of Interstitial Lung Abnormality Patterns with an Ensemble of Deep Convolutional Neural Networks. Sci. Rep. 2020, 10, 338.
- Agarwala, S.; Kale, M.; Kumar, D.; Swaroop, R.; Kumar, A.; Kumar Dhara, A.; Basu Thakur, S.; Sadhu, A.; Nandi, D. Deep Learning for Screening of Interstitial Lung Disease Patterns in High-Resolution CT Images. Clin. Radiol. 2020, 75, 481.e1–481.e8.
- Kim, M.S.; Choe, J.; Hwang, H.J.; Lee, S.M.; Yun, J.; Kim, N.; Ko, M.-S.; Yi, J.; Yu, D.; Seo, J.B. Interstitial Lung Abnormalities (ILA) on Routine Chest CT: Comparison of Radiologists’ Visual Evaluation and Automated Quantification. Eur. J. Radiol. 2022, 157, 110564.
- Nishikiori, H.; Kuronuma, K.; Hirota, K.; Yama, N.; Suzuki, T.; Onodera, M.; Onodera, K.; Ikeda, K.; Mori, Y.; Asai, Y.; et al. Deep-Learning Algorithm to Detect Fibrosing Interstitial Lung Disease on Chest Radiographs. Eur. Respir. J. 2023, 61, 2102269.
- Onishchenko, D.; Marlowe, R.J.; Ngufor, C.G.; Faust, L.J.; Limper, A.H.; Hunninghake, G.M.; Martinez, F.J.; Chattopadhyay, I. Screening for Idiopathic Pulmonary Fibrosis Using Comorbidity Signatures in Electronic Health Records. Nat. Med. 2022, 28, 2107–2116.
- Axelsson, G.T.; Gudmundsson, G.; Pratte, K.A.; Aspelund, T.; Putman, R.K.; Sanders, J.L.; Gudmundsson, E.F.; Hatabu, H.; Gudmundsdottir, V.; Gudjonsson, A.; et al. The Proteomic Profile of Interstitial Lung Abnormalities. Am. J. Respir. Crit. Care Med. 2022, 206, 337–346.
- Pawar, S.P.; Talbar, S.N. Two-Stage Hybrid Approach of Deep Learning Networks for Interstitial Lung Disease Classification. BioMed Res. Int. 2022, 2022, 7340902.
- Huang, S.; Lee, F.; Miao, R.; Si, Q.; Lu, C.; Chen, Q. A Deep Convolutional Neural Network Architecture for Interstitial Lung Disease Pattern Classification. Med. Biol. Eng. Comput. 2020, 58, 725–737.
- Choe, J.; Hwang, H.J.; Seo, J.B.; Lee, S.M.; Yun, J.; Kim, M.-J.; Jeong, J.; Lee, Y.; Jin, K.; Park, R.; et al. Content-Based Image Retrieval by Using Deep Learning for Interstitial Lung Disease Diagnosis with Chest CT. Radiology 2022, 302, 187–197.
- Koo, C.W.; Williams, J.M.; Liu, G.; Panda, A.; Patel, P.P.; Frota Lima, L.M.M.; Karwoski, R.A.; Moua, T.; Larson, N.B.; Bratt, A. Quantitative CT and Machine Learning Classification of Fibrotic Interstitial Lung Diseases. Eur. Radiol. 2022, 32, 8152–8161.
- Furukawa, T.; Oyama, S.; Yokota, H.; Kondoh, Y.; Kataoka, K.; Johkoh, T.; Fukuoka, J.; Hashimoto, N.; Sakamoto, K.; Shiratori, Y.; et al. A Comprehensible Machine Learning Tool to Differentially Diagnose Idiopathic Pulmonary Fibrosis from Other Chronic Interstitial Lung Diseases. Respirology 2022, 27, 739–746.
- Christe, A.; Peters, A.A.; Drakopoulos, D.; Heverhagen, J.T.; Geiser, T.; Stathopoulou, T.; Christodoulidis, S.; Anthimopoulos, M.; Mougiakakou, S.G.; Ebner, L. Computer-Aided Diagnosis of Pulmonary Fibrosis Using Deep Learning and CT Images. Investig. Radiol. 2019, 54, 627–632.
- Bratt, A.; Williams, J.M.; Liu, G.; Panda, A.; Patel, P.P.; Walkoff, L.; Sykes, A.-M.G.; Tandon, Y.K.; Francois, C.J.; Blezek, D.J.; et al. Predicting Usual Interstitial Pneumonia Histopathology from Chest CT Imaging With Deep Learning. Chest 2022, 162, 815–823.
- Yang, F.; Tang, Z.-R.; Chen, J.; Tang, M.; Wang, S.; Qi, W.; Yao, C.; Yu, Y.; Guo, Y.; Yu, Z. Pneumoconiosis Computer Aided Diagnosis System Based on X-Rays and Deep Learning. BMC Med. Imaging 2021, 21, 189.
- Horimasu, Y.; Ohshimo, S.; Yamaguchi, K.; Sakamoto, S.; Masuda, T.; Nakashima, T.; Miyamoto, S.; Iwamoto, H.; Fujitaka, K.; Hamada, H.; et al. A Machine-Learning Based Approach to Quantify Fine Crackles in the Diagnosis of Interstitial Pneumonia: A Proof-of-Concept Study. Medicine 2021, 100, e24738.
- Plantier, L.; Smolinska, A.; Fijten, R.; Flamant, M.; Dallinga, J.; Mercadier, J.J.; Pachen, D.; d’Ortho, M.P.; van Schooten, F.J.; Crestani, B.; et al. The Use of Exhaled Air Analysis in Discriminating Interstitial Lung Diseases: A Pilot Study. Respir. Res. 2022, 23, 12.
- Zhang, H.; Wang, S.; Huang, T. Identification of Chronic Hypersensitivity Pneumonitis Biomarkers with Machine Learning and Differential Co-Expression Analysis. Curr. Gene Ther. 2021, 21, 299–303.
- Li, Z.; Wang, S.; Zhao, H.; Yan, P.; Yuan, H.; Zhao, M.; Wan, R.; Yu, G.; Wang, L. Artificial Neural Network Identified the Significant Genes to Distinguish Idiopathic Pulmonary Fibrosis. Sci. Rep. 2023, 13, 1225.
- Kim, G.H.J.; Weigt, S.S.; Belperio, J.A.; Brown, M.S.; Shi, Y.; Lai, J.H.; Goldin, J.G. Prediction of Idiopathic Pulmonary Fibrosis Progression Using Early Quantitative Changes on CT Imaging for a Short Term of Clinical 18-24-Month Follow-Ups. Eur. Radiol. 2020, 30, 726–734.
- Handa, T.; Tanizawa, K.; Oguma, T.; Uozumi, R.; Watanabe, K.; Tanabe, N.; Niwamoto, T.; Shima, H.; Mori, R.; Nobashi, T.W.; et al. Novel Artificial Intelligence-Based Technology for Chest Computed Tomography Analysis of Idiopathic Pulmonary Fibrosis. Ann. Am. Thorac. Soc. 2022, 19, 399–406.
- Budzikowski, J.D.; Foy, J.J.; Rashid, A.A.; Chung, J.H.; Noth, I.; Armato, S.G. 3rd Radiomics-Based Assessment of Idiopathic Pulmonary Fibrosis Is Associated with Genetic Mutations and Patient Survival. J. Med. Imaging Bellingham 2021, 8, 031903.
- Liang, C.-H.; Liu, Y.-C.; Wan, Y.-L.; Yun, C.-H.; Wu, W.-J.; López-González, R.; Huang, W.-M. Quantification of Cancer-Developing Idiopathic Pulmonary Fibrosis Using Whole-Lung Texture Analysis of HRCT Images. Cancers 2021, 13, 5600.
- Aoki, R.; Iwasawa, T.; Saka, T.; Yamashiro, T.; Utsunomiya, D.; Misumi, T.; Baba, T.; Ogura, T. Effects of Automatic Deep-Learning-Based Lung Analysis on Quantification of Interstitial Lung Disease: Correlation with Pulmonary Function Test Results and Prognosis. Diagnostics 2022, 12, 3038.
- Bowman, W.S.; Newton, C.A.; Linderholm, A.L.; Neely, M.L.; Pugashetti, J.V.; Kaul, B.; Vo, V.; Echt, G.A.; Leon, W.; Shah, R.J.; et al. Proteomic Biomarkers of Progressive Fibrosing Interstitial Lung Disease: A Multicentre Cohort Analysis. Lancet Respir. Med. 2022, 10, 593–602.
- Mayr, C.H.; Simon, L.M.; Leuschner, G.; Ansari, M.; Schniering, J.; Geyer, P.E.; Angelidis, I.; Strunz, M.; Singh, P.; Kneidinger, N.; et al. Integrative Analysis of Cell State Changes in Lung Fibrosis with Peripheral Protein Biomarkers. EMBO Mol. Med. 2021, 13, e12871.
More
Information
Subjects:
Respiratory System
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
609
Revisions:
2 times
(View History)
Update Date:
20 Jul 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




