
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | LAURA LAFON HUGHES | -- | 3636 | 2023-06-26 10:19:32 | | | |
| 2 | Fanny Huang | Meta information modification | 3636 | 2023-06-29 08:08:01 | | | | |
| 3 | Fanny Huang | + 14 word(s) | 3650 | 2023-07-10 03:12:43 | | |
Video Upload Options
SARS-CoV-2 components disturb the transport of certain proteins through the nuclear pores. Some SARS-CoV-2 structural proteins such as Spike (S) and Nucleocapsid (N), most non-structural proteins (remarkably, Nsp1 and Nsp3), as well as some accessory proteins (ORF3d, ORF6, ORF9a) can reach the nucleoplasm either due to their nuclear localization signals (NLS) or taking a shuttle with other proteins. A percentage of SARS-CoV-2 RNA can also reach the nucleoplasm. Remarkably, controversy has recently been raised by proving that-at least under certain conditions-, SARS-CoV-2 sequences can be retrotranscribed and inserted as DNA in the host genome, giving rise to chimeric genes. In turn, the expression of viral-host chimeric proteins could potentially create neo-antigens, activate autoimmunity and promote a chronic pro-inflammatory state.
1. Introduction
| Protein | Blocks TF Activation | Blocks TF Translocation | Interacts with NTRs or Nups | Detected in Nucleus? | Highlights/Comments |
|---|---|---|---|---|---|
| Nsp1 | True No: STAT-P [3] |
NXF1-NXT1 [3] | Yes [4][5] | Reduces host mRNA export [6]: Alters host cell transcriptome [7]: Inhibits HDAC2 transport [3]: Interacts with DNA Pol a [8]. |
|
| Nsp2 | No [4] | ||||
| Nsp3 | True No: NFKB-P No: IRF3-P No: STAT-P [3] |
Yes (NSP3-Nt) No NSP3-Ct [4] |
PLPRO Protease. 3 Macrodomains. Canonical Macrodomain binds PAR. Non-canonical Macrodomains bind G4s. |
||
| Nsp4 | GP210 [9] | No [4] | |||
| Nsp5 | True No: IRF3-P [3] |
IRF3 [6] | Yes [4] | 3-CLPRO, Main Protease. | |
| Nsp6 | No> IRF3-P; STAT-P [6] | Yes [4] | |||
| Nsp7 | Yes [4] | Suppresses IFN-α signaling [3]. | |||
| Nsp8 | True; No>IRF3-P [3] |
No [4] | |||
| Nsp9 | True [3] | Nup54, Nup58,Nup 62, Nup 88, Nup214 [9] Nup 62 [3] | Yes [4] | ||
| Nsp10 | True [3] | Yes [4] | |||
| Nsp11 | |||||
| Nsp12 | True [3] | IRF3 [6] | Yes [4] | RNA-dep RNA-pol. | |
| Nsp13 | No> NF-KB-P; IRF3-P; STAT-P [3][6] | Yes [4] | Colocalizing with SC35 [4]. | ||
| Nsp14 | True [3] | IRF3 [6] | Yes [4] | Exoribonuclease | |
| Nsp15 | True [3] | IRF3 [6] | NTF2 [9] | Yes [4] | RNA endonuclease |
| Nsp16 | True [6] | Yes [4] |
| Protein | Blocks TF Activation | Blocks TF Translocation | Interacts with NTRs or Nups | Detected in Nucleus? | Highlights/Comments |
|---|---|---|---|---|---|
| ORF2. S | No [4] Yes [10][11] |
Has an NLS [11]; Bears a Superantigen motif [12]; Alters cardiomyocyte metabolism and functions [13]; Induces pro-oncogenic cascades [14]; Induces KSHV reactivation [15]; Predicted NES [16]. |
|||
| ORF4.E | Yes [4] | ||||
| ORF5.M | No [4] | ||||
| ORF9a.N | No: IRF3-P; No: STAT-P [6] |
IRF3 STAT1/2 [6] |
No [4] | Biphasic effect on IFN signaling. Low N concentration diminishes it, while high N concentration enhances it and could participate in cytokine storms [9]. Involved in liquid–liquid demixing and IKK sequestration [17]. Localizes to nucleus and nucleolus in IBV and MHV CoV, but not in SARS-CoV (in spite of 3 putative NLS, NoLS and NES). |
| Protein | Blocks TF Activation | Blocks TF Translocation | Interacts with NTRs or Nups | Detected in Nucleus? | Highlights/Comments |
|---|---|---|---|---|---|
| ORF3a | No: STAT-P [6] | No [4] | |||
| ORF3b | True [6] | No [4] | Immunodominant protein. Induces high levels of antibodies [6]. Predicted NES: IITLKKRWQLAL [16] | ||
| ORF6 | True [6] | IRF3. [6] STAT1 through Imp-α1, Impβ1 linkage to ER. Nup-98-RAE1 [9] |
Imp-α1, Impβ1 Nup-98-RAE1 RanBP2/Nup358, Nup160, Nup188, Nup210, Nup 37, Nup93, Imp-5, Imp-8, RanBP6, XPO3, CRM1 [9] |
No. [4] | Alters host mRNA transport [9]; Bidirectional transport disruption. |
| ORF7a | STAT-P [6] | No [4] | |||
| ORF7b | STAT-P [6] | No [4] | |||
| ORF8 | IRF3 [6] | No [4] | NFKB promoter inhibition? | ||
| ORF9b | Small enough to enter through passive diffusion. Interacts with CRM1 exportin [18]. |
No [4] Yes [18] |
Predicted NES [16]; Has an NES [18]; Affect IFN signaling through TOM70 [6]; Triggers apoptosis if retained in the nucleus [18]; Alters cardiomyocytes [13]. |
||
| ORF10 | No [4] | Not essential [6] |
2. SARS-CoV-2 Modulates IFN Signalling
3. SARS-CoV-2 Regulates Nucleocytoplasmic Shuttling
4. SARS-CoV-2 Proteins: Nuclear Localization
5. SARS-CoV-2 RNA: Nuclear Localization
6. Evidence of SARS-CoV-2 sgRNA Retrotranscription and Insertion in the Host Genome
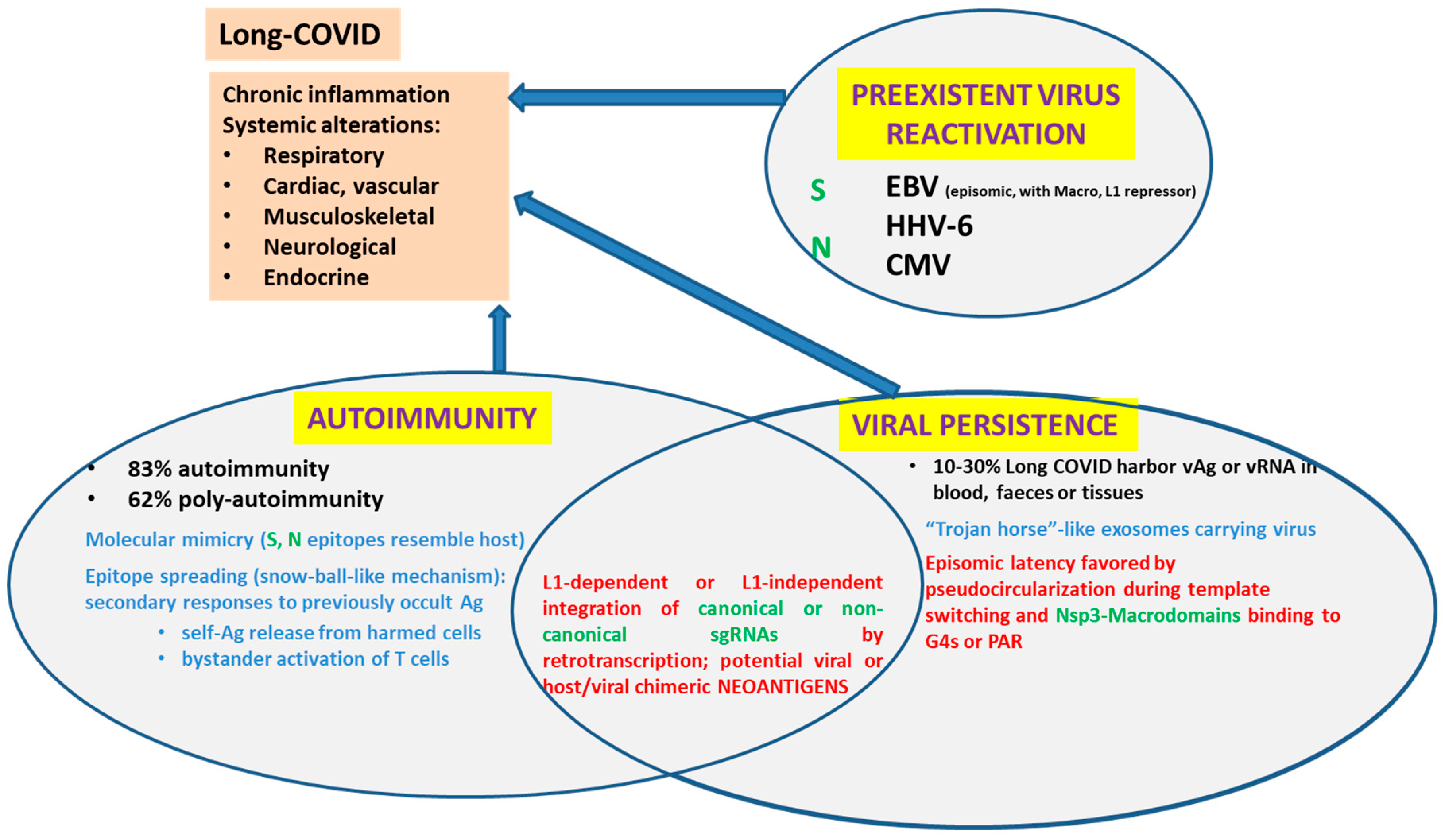
References
- Masters, P.S. The Molecular Biology of Coronaviruses. Adv. Virus Res. 2006, 66, 193–292.
- Brierley, I.; Digard, P.; Inglis, S.C. Characterization of an efficient coronavirus ribosomal frameshifting signal: Requirement for an RNA pseudoknot. Cell 1989, 57, 537–547.
- Low, Z.Y.; Zabidi, N.Z.; Yip, A.J.W.; Puniyamurti, A.; Chow, V.T.K.; Lal, S.K. SARS-CoV-2 Non-Structural Proteins and Their Roles in Host Immune Evasion. Viruses 2022, 14, 1991.
- Zhang, J.; Cruz-cosme, R.; Zhuang, M.-W.; Liu, D.; Liu, Y.; Teng, S.; Wang, P.-H.; Tang, Q.A. A systemic and molecular study of subcellular localization of SARS-CoV-2 proteins. Signal Transduct. Target. Ther. 2020, 5, 269.
- Zhang, K.; Miorin, L.; Makio, T.; Dehghan, I.; Gao, S.; Xie, Y.; Zhong, H.; Esparza, M.; Kehrer, T.; Kumar, A.; et al. Nsp1 protein of SARS-CoV-2 disrupts the mRNA export machinery to inhibit host gene expression. Sci. Adv. 2021, 7, eabe7386.
- Pizzato, M.; Baraldi, C.; Boscato Sopetto, G.; Finozzi, D.; Gentile, C.; Gentile, M.D.; Marconi, R.; Paladino, D.; Raoss, A.; Riedmiller, I.; et al. SARS-CoV-2 and the Host Cell: A Tale of Interactions. Front. Virol. 2022, 1, 815388.
- Kamitani, W.; Huang, C.; Narayanan, K.; Lokugamage, K.G.; Makino, S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009, 16, 1134–1140.
- Rapti, V.; Tsaganos, T.; Vathiotis, I.; Syrigos, N.; Li, P.; Poulakou, G. New Insights into SARS-CoV-2 and Cancer Cross-Talk: Does a Novel Oncogenesis Driver Emerge? Vaccines 2022, 10, 1607.
- Shen, Q.; Wang, Y.E.; Palazzo, A.F. Crosstalk between nucleocytoplasmic trafficking and the innate immune response to viral infection. J. Biol. Chem. 2021, 297, 100856.
- Osan, J.K.; DeMontigny, B.A.; Mehedi, M. Immunohistochemistry for protein detection in PFA-fixed paraffin-embedded SARS-CoV-2-infected COPD airway epithelium. STAR Protoc. 2021, 2, 100663.
- Sattar, S.; Kabat, J.; Jerome, K.; Feldmann, F.; Bailey, K.; Mehedi, M. Nuclear translocation of spike mRNA and protein is a novel feature of SARS-CoV-2. Front. Microbiol. 2023, 14, 1073789.
- Vojdani, A.; Vojdani, E.; Saidara, E.; Maes, M. Persistent SARS-CoV-2 Infection, EBV, HHV-6 and Other Factors May Contribute to Inflammation and Autoimmunity in Long COVID. Viruses 2023, 15, 400.
- Zhang, P.; Liu, Y.; Li, C.; Stine, L.D.; Wang, P.-H.; Turnbull, M.W.; Wu, H.; Liu, Q. Ectopic expression of SARS-CoV-2 S and ORF-9B proteins alters metabolic profiles and impairs contractile function in cardiomyocytes. Front. Cell Dev. Biol. 2023, 11, 1110271.
- Lai, Y.-J.; Chao, C.-H.; Liao, C.-C.; Lee, T.-A.; Hsu, J.-M.; Chou, W.-C.; Wang, J.; Huang, H.-C.; Chang, S.-J.; Lin, Y.-L.; et al. Epithelial-mesenchymal transition induced by SARS-CoV-2 required transcriptional upregulation of Snail. Am. J. Cancer Res. 2021, 11, 2278–2290.
- Chen, J.; Dai, L.; Barrett, L.; James, J.; Plaisance-Bonstaff, K.; Post, S.R.; Qin, Z. SARS-CoV-2 proteins and anti-COVID-19 drugs induce lytic reactivation of an oncogenic virus. Commun. Biol. 2021, 4, 682.
- Kashyap, T.; Murray, J.; Walker, C.J.; Chang, H.; Tamir, S.; Hou, B.; Shacham, S.; Kauffman, M.G.; Tripp, R.A.; Landesman, Y. Selinexor, a novel selective inhibitor of nuclear export, reduces SARS-CoV-2 infection and protects the respiratory system in vivo. Antivir. Res. 2021, 192, 105115.
- Cascarina, S.M.; Ross, E.D. Phase separation by the SARS-CoV-2 nucleocapsid protein: Consensus and open questions. J. Biol. Chem. 2022, 298, 101677.
- Sharma, K.; Åkerström, S.; Sharma, A.K.; Chow, V.T.K.; Teow, S.; Abrenica, B.; Booth, S.A.; Booth, T.F.; Mirazi-mi, A.; Lal, S.K. SARS-CoV 9b protein diffuses into nucleus, undergoes active Crm1 mediated nucleocytoplasmic export and triggers apoptosis when retained in the nucleus. PLoS ONE 2011, 6, e19436.
- Fung, T.S.; Liu, D.X. Human Coronavirus: Host-Pathogen Interaction. Annu. Rev. Microbiol. 2019, 73, 529–557.
- Lei, X.; Dong, X.; Ma, R.; Wang, W.; Xiao, X.; Tian, Z.; Wang, C.; Wang, Y.; Li, L.; Ren, L.; et al. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020, 11, 3810.
- Arora, A.; Kolberg, J.E.; Badarinarayan, S.S.; Munot, D.; Müller, M.; Sauter, D.; Bansal, V. SARS-CoV-2 infection activates endogenous retroviruses of the LTR69 subfamily. Mol. Biol. 2023, preprint.
- Zhao, Y.; Sui, L.; Wu, P.; Wang, W.; Tan, G.; Wang, Z.; Yu, Y.; Hou, Z.; Wang, G.; Liu, Q. SARS-CoV-2 nucleocapsid protein dually regulates innate immune responses. Microbiology 2021, preprint.
- Paci, G.; Caria, J.; Lemke, E.A. Cargo transport through the nuclear pore complex at a glance. J. Cell Sci. 2021, 134, jcs247874.
- Boschi, C.; Scheim, D.E.; Bancod, A.; Militello, M.; Bideau, M.L.; Colson, P.; Fantini, J.; Scola, B.L. SARS-CoV-2 Spike Protein Induces Hemagglutination: Implications for COVID-19 Morbidities and Therapeutics and for Vaccine Adverse Effects. Int. J. Mol. Sci. 2022, 23, 15480.
- Yadav, R.; Chaudhary, J.K.; Jain, N.; Chaudhary, P.K.; Khanra, S.; Dhamija, P.; Sharma, A.; Kumar, A.; Handu, S. Role of Structural and Non-Structural Proteins and Therapeutic Targets of SARS-CoV-2 for COVID-19. Cells 2021, 10, 821.
- Timani, K.A.; Liao, Q.; Ye, L.; Zeng, Y.; Liu, J.; Zheng, Y.; Ye, L.; Yang, X.; Lingbao, K.; Gao, J.; et al. Nuclear/nucleolar localization properties of C-terminal nucleocapsid protein of SARS coronavirus. Virus Res. 2005, 114, 23–34.
- Wu, H.-Y.; Brian, D.A. 5′-Proximal Hot Spot for an Inducible Positive-to-Negative-Strand Template Switch byCoronavirus RNA-Dependent RNA Polymerase. J. Virol. 2007, 81, 3206–3215.
- Wu, C.-H.; Chen, P.-J.; Yeh, S.-H. Nucleocapsid Phosphorylation and RNA Helicase DDX1 Recruitment Enables Coronavirus Transition from Discontinuous to Continuous Transcription. Cell Host Microbe 2014, 16, 462–472.
- Sola, I.; Almazán, F.; Zúñiga, S.; Enjuanes, L. Continuous and Discontinuous RNA Synthesis in Coronaviruses. Annu. Rev. Virol. 2015, 2, 265–288.
- McBride, R.; van Zyl, M.; Fielding, B. The Coronavirus Nucleocapsid Is a Multifunctional Protein. Viruses 2014, 6, 2991–3018.
- Wulan, W.N.; Heydet, D.; Walker, E.J.; Gahan, M.E.; Ghildyal, R. Nucleocytoplasmic transport of nucleocapsid proteins of enveloped RNA viruses. Front. Microbiol. Sec. Virol. 2015, 6, 553.
- Andersen, K.G.; Rambaut, A.; Lipkin, W.I.; Holmes, E.C.; Garry, R.F. The proximal origin of SARS-CoV-2. Nat. Med. 2020, 26, 450–452.
- Pradhan, P.; Pandey, A.K.; Mishra, A.; Gupta, P.; Tripathi, P.K.; Menon, M.B.; Gomes, J.; Vivekanandan, P.; Kundu, B. Uncanny similarity of unique inserts in the 2019-nCoV spike protein to HIV-1 gp120 and Gag. Evol. Biol. 2020; withdrawn preprint.
- Zhang, C.; Zheng, W.; Huang, X.; Bell, E.W.; Zhou, X.; Zhang, Y. Protein Structure and Sequence Reanalysis of 2019-nCoV Genome Refutes Snakes as Its Intermediate Host and the Unique Similarity between Its Spike Protein Insertions and HIV-1. J. Proteome Res. 2020, 19, 1351–1360.
- Feng, Z.; Diao, B.; Wang, R.; Wang, G.; Wang, C.; Tan, Y.; Liu, L.; Wang, C.; Liu, Y.; Liu, Y.; et al. The Novel Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) Directly Decimates Human Spleens and Lymph Nodes. Infect. Dis. (Except. HIV/AIDS), 2020; preprint.
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients with Coronavirus Disease 2019 (COVID-19). Infect. Dis. (Except. HIV/AIDS), 2020; preprint.
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134.
- Braun, E.; Sauter, D. Furin-mediated protein processing in infectious diseases and cancer. Clin. Transl. Immunol. 2019, 8, e1073.
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. 2019, 38, 173.
- AFehr, R.; Jankevicius, G.; Ahel, I.; Perlman, S. Viral Macrodomains: Unique Mediators of Viral Replication and Pathogenesis. Trends Microbiol. 2018, 26, 598–610.
- Hottiger, M.O. SnapShot: ADP-Ribosylation Signaling. Mol. Cell 2015, 58, 1134.
- Egloff, M.-P.; Malet, H.; Putics, Á.; Heinonen, M.; Dutartre, H.; Frangeul, A.; Gruez, A.; Campanacci, V.; Cam-billau, C.; Ziebuhr, J.; et al. Structural and Functional Basis for ADP-Ribose and Poly(ADP-Ribose) Binding by Viral Macro Domains. J. Virol. 2006, 80, 8493–8502.
- Tan, J.; Vonrhein, C.; Smart, O.S.; Bricogne, G.; Bollati, M.; Kusov, Y.; Hansen, G.; Mesters, J.R.; Schmidt, C.L.; Hilgenfeld, R. The SARS-Unique Domain (SUD) of SARS Coronavirus Contains Two Macrodomains That Bind G-Quadruplexes. PLoS Pathog. 2009, 5, e1000428.
- Kusov, Y.; Tan, J.; Alvarez, E.; Enjuanes, L.; Hilgenfeld, R. A G-quadruplex-binding macrodomain within the ‘SARS-unique domain’ is essential for the activity of the SARS-coronavirus replication–transcription complex. Virology 2015, 484, 313–322.
- Ruggiero, E.; Richter, S.N. G-quadruplexes and G-quadruplex ligands: Targets and tools in antiviral therapy. Nucleic Acids Res. 2018, 46, 3270–3283.
- Lieberman, P.M. Epigenetics and Genetics of Viral Latency. Cell Host Microbe 2016, 19, 619–628.
- Lee, J.Y.; Wing, P.A.; Gala, D.S.; Noerenberg, M.; Järvelin, A.I.; Titlow, J.; Zhuang, X.; Palmalux, N.; Iselin, L.; Thompson, M.K.; et al. Absolute quantitation of individual SARS-CoV-2 RNA molecules provides a new paradigm for infection dynamics and variant differences. eLife 2022, 11, e74153.
- Stueland, M.; Wang, T.; Park, H.Y.; Mili, S. RDI Calculator: An Analysis Tool to Assess RNA Distributions in Cells. Sci. Rep. 2019, 9, 8267.
- Zhang, L.; Richards, A.; Barrasa, M.I.; Hughes, S.H.; Young, R.A.; Jaenisch, R. Reverse-transcribed SARS-CoV-2 RNA can integrate into the genome of cultured human cells and can be expressed in patient-derived tissues. Proc. Natl. Acad. Sci. USA 2021, 118, e2105968118.
- Yan, B.; Chakravorty, S.; Mirabelli, C.; Wang, L.; Trujillo-Ochoa, J.L.; Chauss, D.; Kumar, D.; Lionakis, M.S.; Olson, M.R.; Wobus, C.E.; et al. Host-Virus Chimeric Events in SARS-CoV-2-Infected Cells Are Infrequent and Artifactual. J. Virol. 2021, 95, e00294-21.
- Smits, N.; Rasmussen, J.; Bodea, G.O.; Amarilla, A.A.; Gerdes, P.; Sanchez-Luque, F.J.; Ajjikuttira, P.; Modhiran, N.; Liang, B.; Faivre, J.; et al. No evidence of human genome integration of SARS-CoV-2 found by long-read DNA sequencing. Cell Rep. 2021, 36, 109530.
- Podlaha, O.; Wu, G.; Downie, B.; Ramamurthy, R.; Gaggar, A.; Subramanian, M.; Ye, Z.; Jiang, Z. Genomic modeling of hepatitis B virus integration frequency in the human genome. PLoS ONE 2019, 14, e0220376.




