Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jovanka Gencel-Augusto | -- | 2399 | 2023-06-23 00:13:35 | | | |
| 2 | Dean Liu | -69 word(s) | 2330 | 2023-06-25 04:29:27 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Gencel-Augusto, J.; Wu, W.; Bivona, T.G. Long Non-Coding RNAs as Emerging Targets. Encyclopedia. Available online: https://encyclopedia.pub/entry/45980 (accessed on 07 February 2026).
Gencel-Augusto J, Wu W, Bivona TG. Long Non-Coding RNAs as Emerging Targets. Encyclopedia. Available at: https://encyclopedia.pub/entry/45980. Accessed February 07, 2026.
Gencel-Augusto, Jovanka, Wei Wu, Trever G. Bivona. "Long Non-Coding RNAs as Emerging Targets" Encyclopedia, https://encyclopedia.pub/entry/45980 (accessed February 07, 2026).
Gencel-Augusto, J., Wu, W., & Bivona, T.G. (2023, June 22). Long Non-Coding RNAs as Emerging Targets. In Encyclopedia. https://encyclopedia.pub/entry/45980
Gencel-Augusto, Jovanka, et al. "Long Non-Coding RNAs as Emerging Targets." Encyclopedia. Web. 22 June, 2023.
Copy Citation
Long non-coding RNAs (LncRNAs) are non-protein coding molecules longer than 200 nucleotides. They play essential roles in normal cell function and development, and can contribute to diseases such as cancer when dysregulated. Although lncRNAs have oncogenic or tumor-suppressive properties in lung cancer and can serve as stable biomarkers, this is still an understudied field.
LncRNAs
lung cancer
metastasis
therapy resistance
biomarkers
1. Introduction
Long non-coding RNAs (LncRNAs) are broadly defined as RNAs that usually do not encode for proteins and that are longer than 200 nucleotides. These are messenger RNA (mRNA)-like molecules that are transcribed by polymerase II, 5′ capped, and have a 3′ poly-A tail. Because they can form complex secondary structures, they often have functions. Many lncRNAs are preferentially found in the nucleus, where they participate in the regulation of chromatin organization and transcription, often by forming lncRNA-DNA triplex [1][2][3], as well as through the formation of nuclear speckles and regulation of splicing. In the cytoplasm, lncRNAs can regulate mRNA stability, bind to other non-coding RNAs, and modulate protein post-translational modifications and protein function [4][5][6][7][8].
LncRNAs have been studied in mammals since the early 1990s due to their involvement in developmental processes. For example, the Xist (X-inactive specific transcript) lncRNA contributes to reshaping the architecture of chromatin to achieve X chromosome-silencing in early embryonic development [9]. The H19 lncRNA is involved in genomic imprinting and regulation of the insulin growth factor 2 (IGF2) and other genes involved in embryonic growth [10][11]. The study of HOX genes, master regulators of embryonic development, led to the discovery of the lncRNA HOTAIR (Homeobox transcript antisense intergenic RNA). HOTAIR is transcribed from the antisense strand of the HOXC gene.
HOTAIR has been reported to repress the transcription of the HOXD loci via interaction with PRC2 (polycomb repressive complex 2) [12], although in vivo models developed later report conflicting results regarding HOXC or HOXD genes regulation by HOTAIR [13][14]. Because of the importance of physiological context to understand lncRNA molecular function, more recently, other groups have reported exclusively in vivo approaches using animal models of lncRNA genetic ablation. For example, a comprehensive study developed 18 knock-out (KO) mouse models for less well-known lncRNAs with human orthologs. Certain lncRNAs’ expression was highly tissue-specific (such as Fendrr, Manr and linc–Cox2 expressed mainly in lung), supporting a unique physiological role, while other lncRNAs were more ubiquitously expressed. Three of the analyzed lncRNAs were required for embryonic development (Fendrr, Mdgt, Peril), speaking to their fundamental functions [15]. Thus, the involvement of lncRNAs in normal cell physiology and organism development suggests that these may also control disease-related processes such as cancer.
Lung cancer is the leading cause of cancer-related mortality in the U.S., and non-small cell lung cancer (NSCLC) is the most common subtype. Lung cancer is commonly diagnosed in late stages, where patients present distant metastasis with 9% having a 5-year survival rate [16]. Although the development of targeted therapies (e.g., tyrosine kinase inhibitors, TKIs) has improved patient outcomes, their clinical efficacy is often limited by both innate and acquired resistance, permitting tumor progression and recurrence leading to poor survival rates [17]. It is imperative to better understand molecular drivers of tumorigenesis, metastasis, and therapy resistance in lung cancer to develop improved therapeutic strategies. LncRNAs are emerging as important molecules in cancer due to their oncogenic or tumor-suppressive functions.
2. Role of lncRNAs in Lung Cancer
Because of the role of lncRNAs in regulating a diverse array of cellular functions, deregulation of their activities is involved in cancer. Some mechanisms reported for lncRNAs functions in cancer are as follows: acting as miRNA sponges to modulate activity on their targets; interacting with histone-modifier enzymes to modulate known oncogene/tumor suppressor gene expression; interacting with transcription factors to repress/activate their transcriptional programs; acting as anti-sense molecules for tumor suppressor mRNAs, among other mechanisms [2][18]. Importantly, regulation of lncRNA expression and function in cancer follows similar principles to that of known oncogenes and tumor suppressors, as it can be mediated by DNA methylation [19], amplification or deletion [20], and mutation or SNPs of DNA sequences [21][22]. Researchers provide examples of well-studied lncRNAs that are reported to have oncogenic or tumor-suppressive roles in lung cancer, as well as those with controversial functions.
MALAT1 (Metastasis-Associated Lung Adenocarcinoma Transcript 1) was one of the first lncRNAs described to be associated with cancer. In 2003, Ji et al. analyzed the gene expression profile in human primary lung cancer tumors that subsequently metastasized or those that did not metastasize and compared their transcriptional signatures. They identified a Metastasis-Associated Lung Adenocarcinoma Transcript 1, named MALAT1, for its higher expression in primary tumors that metastasized. They also found a significant correlation between higher level of MALAT1 expression in stage I lung cancer and worse survival outcomes [23]. Several studies since then have described MALAT1 function in normal physiology and cancer [24][25][26]. Three independent groups developed an in vivo approach to describing MALAT1 physiological function. They found that Malat1 is highly abundant in several mouse tissues and highly conserved across species. Genetic perturbation of the Malat1 locus in mice (using genetic deletion [27][28] or genetic inactivation approaches [29]) did not alter animal development, nuclear speckle formation, splicing, or mRNA stability. However, they described a role for Malat1 in controlling neighboring genes expression in a tissue-specific manner that was not consistent between the three studies, especially that of Neat1, another lncRNA [28][29]. In the context of lung cancer, MALAT1-silencing did not show effects on lung cancer cell proliferation or viability in vitro [27]. To further understand the role of MALAT1 in lung cancer metastasis, Gutschner et al. implanted EBC-1 lung cancer cells into nude mice and treated them with subcutaneous administration of an anti-sense oligonucleotide (ASO) targeting MALAT1. After five weeks of treatment, all primary tumors were excised. Metastasis nodules were analyzed at 12 weeks, indicating fewer and smaller metastatic nodules in the treated group, suggesting a role for MALAT1 in promoting metastasis. Through additional in vitro studies, they report MALAT1 inhibition results in aberrant expression of metastasis-associated genes in cell lines [30]. Although these studies support a role for MALAT1 in promoting metastasis and regulating certain genes expression, other evidence exists for different roles. Kim et al. describe an elegant study that challenges previously reported roles for MALAT1 in metastasis. Using the Malat1 knock-out (KO) mouse model developed by Nakagawa et al. (LacZ and Poly-A sequences were used as transcriptional terminators inserted 69 bp downstream of the transcription start site of Malat1 without deletion of the DNA sequence), Kim et al. crossed these mice with a breast cancer model driven by MMTV-PyMT that mimics human disease. Surprisingly, they found a 7.2-fold increase in metastatic foci and 31-fold increase in the percent of lung areas with metastatic lesions in Malat1-KO mice as compared to Malat1 WT mice, suggesting a role for Malat1 in suppressing breast cancer metastasis to the lung. This phenotype was rescued by transgenic expression of Malat1, suggesting that the RNA product itself diminished metastasis. Additionally, they identified Malat1 interaction with TEAD (transcriptional enhanced associate domain) proteins in mouse-derived tumors and cell lines, which suppressed TEAD-YAP interaction and, therefore, inhibited their pro-metastatic transcriptional program [31]. Of note, DNA elements themselves within a lncRNA locus may be responsible for regulatory functions that are independent from transcript function [32][33][34][35]. In summary, MALAT1 promotes metastasis in lung cancer, but may show opposite functions in different types of cancer depending on cellular context.
The GAS5 (growth arrest-specific 5) gene was first described as a G0-specific gene that is inhibited by serum and growth factors [36]. In vivo, Gas5 genetic deletion (Gas5+/−) in mice decreased bone mass and impaired bone repair, leading to osteoporosis. Mechanistically, Gas5 positively influenced proper cell differentiation through interaction with UPF1 (a DNA/RNA helicase) to accelerate SMAD7 mRNA decay [37]. In lung cancer, GAS5 was found down-regulated in 72 NSCLC tumor samples as compared to their paired adjacent normal tissues, suggesting a tumor-suppressive role. Additionally, low GAS5 expression was correlated with larger tumor size, lower differentiation levels, and higher staging of tumor–node metastasis [38]. A xenograft model of GAS5 overexpression (OE) showed that GAS5 OE markedly decreased tumor size compared with control [38]. Although researchers did not explore a mechanism for GAS5-mediated tumor suppression, other studies implicate a role for GAS5 as a miRNA sponge to negatively influence cell cycle activator genes [39] or positively influence PTEN levels [40]. Additionally, a recent study revealed GAS5 is partially localized to the mitochondria where it modulates energy homeostasis by promoting the de-acetylation of malate dehydrogenase, suppressing breast cancer [41]. Taken together, a role for GAS5 in halting the cell cycle as well as promoting cell differentiation in normal cells supports its tumor-suppressive role reported in lung cancer. Besides GAS5, other lncRNAs have been studied for their tumor-suppressive functions, such as MEG3 and TUG1, reviewed elsewhere [42].
LUCAT1 (Lung Cancer-Associated Transcript 1), first identified as smoke-induced and cancer-associated lncRNA1 (SCAL1) [43], has higher expression in lung cancer as compared to normal controls, and is also found to be overexpressed in several cancer types [44]. A LUCAT1-deficient mouse model has not been reported. Additionally, patients with tumors that express high levels of LUCAT1 showed poorer overall survival as compared to those with lower LUCAT1 expression. Moreover, high LUCAT1 levels were associated with late staging in tumor–lymph node metastasis and higher tumor volume. In NSCLC cell lines A549 and SPC-A1, LUCAT1 modulates p21 and p57 expression by promoting loci methylation through PRC2 [45].
More recently, a role for LUCAT1 in regulating immune responses has been described. LUCAT1 genetic deletion in myeloid cells is found to enhance interferon-mediated gene transcription. LUCAT1 acts as an immune suppressor by interacting with STAT1 and chromatin in the nucleus. It may also act by inhibiting NF-kB functions [46]. These findings suggest a tumor-promoting role for LUCAT1 that is tumor-cell-intrinsic, in addition to a potential non-cell autonomous mechanism via the inhibition of immune surveillance, although this mechanism remains to be explored in the lung cancer context.
HOTAIR has been vastly studied in cancer contexts [47][48]. In NSCLC, tumor samples and cell lines expressed higher HOTAIR levels as compared to normal counterparts [49]. Additionally, high HOTAIR levels correlated with higher tumor grade and presence of lymph node metastases [49][50]. In vitro, HOTAIR has been reported as a direct target of the hypoxia-inducible-factor-1α (HIF-1α), therefore enhancing A549 NSCLC cells’ proliferation, migration, and invasion [51]. In vivo, tail vein injections of SPC-A1 cells with or without siRNA targeting HOTAIR showed that the knock-down condition reduced the number of metastasis nodules found in the lungs of immunocompromised mice [49]. HOTAIR-silencing resulted in a decrease in matrix metalloproteinases (MMPs, which promote invasion and migration) expression and an increase in HOXA5 levels (a tumor suppressor) in cell lines, suggesting HOTAIR acts through the regulation of expression of cancer-related genes [49]. Although this xenograft assay does not account for all steps required for a tumor cell to achieve metastatic colonization (extravasation, survival in blood, seeding of new site, proliferation in new site), and they measured colonization of lungs using a lung cancer cell line (same tissue), it raises the possibility that HOTAIR may be involved in the seeding and survival of cancer cells. Additionally, the absence of a competent immune system challenges interpretation of these results. Development of a HOTAIR transgenic mouse model to understand in vivo implications in lung cancer initiation and progression is necessary, similar to a HOTAIR inducible system recently reported for breast cancer [52]. This model showed that sustained HOTAIR overexpression promotes breast cancer metastasis to lungs. Overall, with the data available, HOTAIR seems to play an oncogenic role in lung cancer; however, robust mechanisms through which this lncRNA function remain to be uncovered.
Most studies focus on the contribution of a single lncRNA to cancer phenotypes. However, whether the lncRNAs described above are expressed simultaneously in tumors with unique or redundant functions remains to be explored in depth. For example, Esposito et al. showed that at least 80 oncogenic lncRNAs are active in NSCLC through a lncRNA-focused CRISPR screen. By further dissecting the role of two candidate lncRNAs, CHiLL1 and GCAWKR, they showed these have distinct cellular localization and non-overlapping targets. Importantly, ASOs targeting both these lncRNAs yielded additive effects, suggesting that they have cooperating functions in NSCLC progression [53]. LncRNAs are generally expressed at lower levels than protein-coding genes [54]. Because of this, researchers speculate that lncRNAs with redundant functions may be expressed simultaneously to compensate for a higher expression of their targets in disease conditions. By examining available TCGA lung adenocarcinoma datasets containing mRNA expression data, researchers did not find significant correlations (negative or positive) among the expression of lncRNAs described here. However, such an analysis in combination with functional studies could shed light on mutual exclusivity relationships between certain lncRNAs. Additionally, whether certain lncRNAs are predominantly expressed at different stages of tumor progression remains to be explored. A new online resource, lncRNAfunc https://ccsm.uth.edu/lncRNAfunc (accessed on 7 May 2023), provides insights on differentially expressed lncRNAs across different cancer types and stages available in TCGA, as well as functional predictions [55]. Although this analysis did not detect any correlations between the lncRNAs mentioned here and lung cancer stages, possibly due to lack of sufficient sample sizes, these lncRNAs did show correlation with stage in other cancers; for example, LUCAT1 was correlated with cancer stage in kidney cancer.
lncRNAs have oncogenic and tumor-suppressive roles in lung cancer, illustrated in Figure 1. LncRNAs interact with protein-coding molecules, resulting in the activation or inactivation of specific signaling pathways in cancer cells. Researchers speculate more lung-cancer-specific lncRNAs will be identified with genome-wide transcriptomic studies.
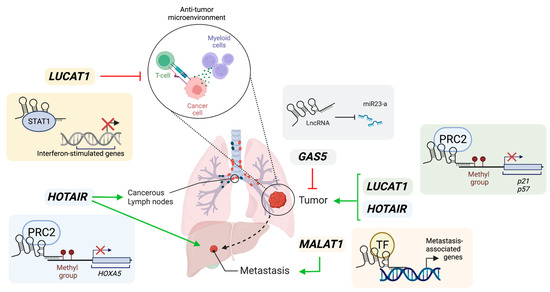
Figure 1. Illustration of roles of lncRNAs in lung cancer. GAS5 acts as a miRNA sponge to activate their mRNA targets and inhibit cancer cell proliferation, therefore acting as a tumor suppressor. LUCAT1 can promote cancer cell proliferation via epigenetic silencing of p21 and p57 loci. Additionally, LUCAT1 may create a pro-tumorigenic microenvironment by inhibiting interferon-mediated responses through the sequestration of STAT1. HOTAIR can promote cancer cell proliferation and metastasis via the recruitment of PRC2 to methylate loci and repress gene expression, such as HOXA5 which is a tumor suppressor. MALAT1 stimulates lung cancer metastasis by potential recruitment of transcription factors (TF) to promoters of metastasis-associated genes. Green arrows represent positive influence. Red arrows represent inhibition. Created with Biorender.com (accessed on 4 June 2023).
References
- Leisegang, M.S.; Bains, J.K.; Seredinski, S.; Oo, J.A.; Krause, N.M.; Kuo, C.C.; Gunther, S.; Sentruk Cetin, N.; Warwick, T.; Cao, C.; et al. HIF1alpha-AS1 is a DNA:DNA:RNA triplex-forming lncRNA interacting with the HUSH complex. Nat. Commun. 2022, 13, 6563.
- O’Leary, V.B.; Ovsepian, S.V.; Carrascosa, L.G.; Buske, F.A.; Radulovic, V.; Niyazi, M.; Moertl, S.; Trau, M.; Atkinson, M.J.; Anastasov, N. PARTICLE, a Triplex-Forming Long ncRNA, Regulates Locus-Specific Methylation in Response to Low-Dose Irradiation. Cell Rep. 2015, 11, 474–485.
- Rakheja, I.; Ansari, A.H.; Ray, A.; Chandra Joshi, D.; Maiti, S. Small molecule quercetin binds MALAT1 triplex and modulates its cellular function. Mol. Ther. Nucleic Acids 2022, 30, 241–256.
- Kung, J.T.; Colognori, D.; Lee, J. Long noncoding RNAs: Past, present, and future. Genetics 2013, 193, 651–669.
- Yao, R.W.; Wang, Y.; Chen, L.L. Cellular functions of long noncoding RNAs. Nat. Cell Biol. 2019, 21, 542–551.
- Guo, C.J.; Xu, G.; Chen, L.L. Mechanisms of Long Noncoding RNA Nuclear Retention. Trends Biochem. Sci. 2020, 45, 947–960.
- Chen, B.; Dragomir, M.P.; Yang, C.; Li, Q.; Horst, D.; Calin, G.A. Targeting non-coding RNAs to overcome cancer therapy resistance. Signal Transduct. Target. Ther. 2022, 7, 121.
- Long, Y.; Wang, X.; Youmans, D.T.; Cech, T.R. How do lncRNAs regulate transcription? Sci. Adv. 2017, 3, eaao2110.
- Wang, W.; Min, L.; Qiu, X.; Wu, X.; Liu, C.; Ma, J.; Zhang, D.; Zhu, L. Biological Function of Long Non-coding RNA (LncRNA) Xist. Front. Cell Dev. Biol. 2021, 9, 645647.
- Monnier, P.; Martinet, C.; Pontis, J.; Stancheva, I.; Ait-Si-Ali, S.; Dandolo, L. H19 lncRNA controls gene expression of the Imprinted Gene Network by recruiting MBD. Proc. Natl. Acad. Sci. USA 2013, 110, 20693–20698.
- Ripoche, M.A.; Kress, C.; Poirier, F.; Dandolo, L. Deletion of the H19 transcription unit reveals the existence of a putative imprinting control element. Genes Dev. 1997, 11, 1596–1604.
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323.
- Li, L.; Liu, B.; Wapinski, O.L.; Tsai, M.C.; Qu, K.; Zhang, J.; Carlson, J.C.; Lin, M.; Fang, F.; Gupta, R.A.; et al. Targeted disruption of Hotair leads to homeotic transformation and gene derepression. Cell Rep. 2013, 5, 3–12.
- Amandio, A.R.; Necsulea, A.; Joye, E.; Mascrez, B.; Duboule, D. Hotair Is Dispensible for Mouse Development. PLoS Genet. 2016, 12, e1006232.
- Sauvageau, M.; Goff, L.A.; Lodato, S.; Bonev, B.; Groff, A.F.; Gerhardinger, C.; Sanchez-Gomez, D.B.; Hacisuleyman, E.; Li, E.; Spence, M.; et al. Multiple knockout mouse models reveal lincRNAs are required for life and brain development. Elife 2013, 2, e01749.
- Cronin, K.A.; Lake, A.J.; Scott, S.; Sherman, R.L.; Noone, A.M.; Howlader, N.; Henley, S.J.; Anderson, R.N.; Firth, A.U.; Ma, J.; et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer 2018, 124, 2785–2800.
- Sabnis, A.J.; Bivona, T.G. Principles of Resistance to Targeted Cancer Therapy: Lessons from Basic and Translational Cancer Biology. Trends Mol. Med. 2019, 25, 185–197.
- Zhang, X.Z.; Liu, H.; Chen, S.R. Mechanisms of Long Non-Coding RNAs in Cancers and Their Dynamic Regulations. Cancers 2020, 12, 1245.
- Shen, S.; Chen, J.; Li, H.; Jiang, Y.; Wei, Y.; Zhang, R.; Zhao, Y.; Chen, F. Large-scale integration of the non-coding RNAs with DNA methylation in human cancers. Cell Rep. 2023, 42, 112261.
- Aprile, M.; Katopodi, V.; Leucci, E.; Costa, V. LncRNAs in Cancer: From garbage to Junk. Cancers 2020, 12, 3220.
- Tong, G.; Tong, W.; He, R.; Cui, Z.; Li, S.; Zhou, B.; Yin, Z. MALAT1 Polymorphisms and Lung Cancer Susceptibility in a Chinese Northeast Han Population. Int. J. Med. Sci. 2022, 19, 1300–1306.
- Ren, M.M.; Xu, S.; Wei, Y.B.; Yang, J.J.; Yang, Y.N.; Sun, S.S.; Li, Y.J.; Wang, P.Y.; Xie, S.Y. Roles of HOTAIR in lung cancer susceptibility and prognosis. Mol. Genet. Genom. Med. 2020, 8, e1299.
- Ji, P.; Diederichs, S.; Wang, W.; Boing, S.; Metzger, R.; Schneider, P.M.; Tidow, N.; Brandt, B.; Buerger, H.; Bulk, E.; et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 2003, 22, 8031–8041.
- Sun, Y.; Ma, L. New Insights into Long Non-Coding RNA MALAT1 in Cancer and Metastasis. Cancers 2019, 11, 216.
- Hou, J.; Zhang, G.; Wang, X.; Wang, Y.; Wang, K. Functions and mechanisms of lncRNA MALAT1 in cancer chemotherapy resistance. Biomark. Res. 2023, 11, 23.
- Jiang, L.; Li, Z.; Wang, R. Long non-coding RNAs in lung cancer: Regulation patterns, biologic function and diagnosis implications (Review). Int. J. Oncol. 2019, 55, 585–596.
- Eissmann, M.; Gutschner, T.; Hammerle, M.; Gunther, S.; Caudron-Herger, M.; Gross, M.; Schirmacher, P.; Rippe, K.; Braun, T.; Zornig, M.; et al. Loss of the abundant nuclear non-coding RNA MALAT1 is compatible with life and development. RNA Biol. 2012, 9, 1076–1087.
- Zhang, B.; Arun, G.; Mao, Y.S.; Lazar, Z.; Hung, G.; Bhattacharjee, G.; Xiao, X.; Booth, C.J.; Wu, J.; Zhang, C.; et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012, 2, 111–123.
- Nakagawa, S.; Ip, J.Y.; Shioi, G.; Tripathi, V.; Zong, X.; Hirose, T.; Prasanth, K.V. Malat1 is not an essential component of nuclear speckles in mice. RNA 2012, 18, 1487–1499.
- Gutschner, T.; Hammerle, M.; Eissmann, M.; Hsu, J.; Kim, Y.; Hung, G.; Revenko, A.; Arun, G.; Stentrup, M.; Gross, M.; et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013, 73, 1180–1189.
- Kim, J.; Piao, H.L.; Kim, B.J.; Yao, F.; Han, Z.; Wang, Y.; Xiao, Z.; Siverly, A.N.; Lawhon, S.E.; Ton, B.N.; et al. Long noncoding RNA MALAT1 suppresses breast cancer metastasis. Nat. Genet. 2018, 50, 1705–1715.
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436.
- Cho, S.W.; Xu, J.; Sun, R.; Mumbach, M.R.; Carter, A.C.; Chen, Y.G.; Yost, K.E.; Kim, J.; He, J.; Nevins, S.A.; et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 2018, 173, 1398–1412.e22.
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455.
- Nunez-Martinez, H.N.; Recillas-Targa, F. Emerging Functions of lncRNA Loci beyond the Transcript Itself. Int. J. Mol. Sci. 2022, 23, 6258.
- Schneider, C.; King, R.M.; Philipson, L. Genes specifically expressed at growth arrest of mammalian cells. Cell 1988, 54, 787–793.
- Li, M.; Xie, Z.; Li, J.; Lin, J.; Zheng, G.; Liu, W.; Tang, S.; Cen, S.; Ye, G.; Li, Z.; et al. GAS5 protects against osteoporosis by targeting UPF1/SMAD7 axis in osteoblast differentiation. Elife 2020, 9, e59079.
- Dong, S.; Qu, X.; Li, W.; Zhong, X.; Li, P.; Yang, S.; Chen, X.; Shao, M.; Zhang, L. The long non-coding RNA, GAS5, enhances gefitinib-induced cell death in innate EGFR tyrosine kinase inhibitor-resistant lung adenocarcinoma cells with wide-type EGFR via downregulation of the IGF-1R expression. J. Hematol. Oncol. 2015, 8, 43.
- Mei, Y.; Si, J.; Wang, Y.; Huang, Z.; Zhu, H.; Feng, S.; Wu, X.; Wu, L. Long Noncoding RNA GAS5 Suppresses Tumorigenesis by Inhibiting miR-23a Expression in Non-Small Cell Lung Cancer. Oncol. Res. 2017, 25, 1027–1037.
- Guo, C.; Song, W.Q.; Sun, P.; Jin, L.; Dai, H.Y. LncRNA-GAS5 induces PTEN expression through inhibiting miR-103 in endometrial cancer cells. J. Biomed. Sci. 2015, 22, 100.
- Sang, L.; Ju, H.Q.; Yang, Z.; Ge, Q.; Zhang, Z.; Liu, F.; Yang, L.; Gong, H.; Shi, C.; Qu, L.; et al. Mitochondrial long non-coding RNA GAS5 tunes TCA metabolism in response to nutrient stress. Nat. Metab. 2021, 3, 90–106.
- Ye, R.; Tang, R.; Gan, S.; Li, R.; Cheng, Y.; Guo, L.; Zeng, C.; Sun, Y. New insights into long non-coding RNAs in non-small cell lung cancer. Biomed. Pharmacother. 2020, 131, 110775.
- Thai, P.; Statt, S.; Chen, C.; Liang, E.; Campbell, C.; Wu, R. Characterization of a novel long noncoding RNA, SCAL1, induced by cigarette smoke and elevated in lung cancer cell lines. Am. J. Respir. Cell Mol. Biol. 2013, 49, 204–211.
- Xing, C.; Sun, S.G.; Yue, Z.Q.; Bai, F. Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother. 2021, 134, 111158.
- Sun, Y.; Jin, S.D.; Zhu, Q.; Han, L.; Feng, J.; Lu, X.Y.; Wang, W.; Wang, F.; Guo, R.H. Long non-coding RNA LUCAT1 is associated with poor prognosis in human non-small lung cancer and regulates cell proliferation via epigenetically repressing p21 and p57 expression. Oncotarget 2017, 8, 28297–28311.
- Agarwal, S.; Vierbuchen, T.; Ghosh, S.; Chan, J.; Jiang, Z.; Kandasamy, R.K.; Ricci, E.; Fitzgerald, K.A. The long non-coding RNA LUCAT1 is a negative feedback regulator of interferon responses in humans. Nat. Commun. 2020, 11, 6348.
- Mahpour, A.; Mullen, A.C. Our emerging understanding of the roles of long non-coding RNAs in normal liver function, disease, and malignancy. JHEP Rep. 2021, 3, 100177.
- Zhu, C.; Wang, X.; Wang, Y.; Wang, K. Functions and underlying mechanisms of lncRNA HOTAIR in cancer chemotherapy resistance. Cell Death Discov. 2022, 8, 383.
- Liu, X.H.; Liu, Z.L.; Sun, M.; Liu, J.; Wang, Z.X.; De, W. The long non-coding RNA HOTAIR indicates a poor prognosis and promotes metastasis in non-small cell lung cancer. BMC Cancer 2013, 13, 464.
- Liu, M.Y.; Li, X.Q.; Gao, T.H.; Cui, Y.; Ma, N.; Zhou, Y.; Zhang, G.J. Elevated HOTAIR expression associated with cisplatin resistance in non-small cell lung cancer patients. J. Thorac. Dis. 2016, 8, 3314–3322.
- Zhou, C.; Ye, L.; Jiang, C.; Bai, J.; Chi, Y.; Zhang, H. Long noncoding RNA HOTAIR, a hypoxia-inducible factor-1alpha activated driver of malignancy, enhances hypoxic cancer cell proliferation, migration, and invasion in non-small cell lung cancer. Tumour Biol. 2015, 36, 9179–9188.
- Ma, Q.; Yang, L.; Tolentino, K.; Wang, G.; Zhao, Y.; Litzenburger, U.M.; Shi, Q.; Zhu, L.; Yang, C.; Jiao, H.; et al. Inducible lncRNA transgenic mice reveal continual role of HOTAIR in promoting breast cancer metastasis. Elife 2022, 11, e79126.
- Esposito, R.; Polidori, T.; Meise, D.F.; Pulido-Quetglas, C.; Chouvardas, P.; Forster, S.; Schaerer, P.; Kobel, A.; Schlatter, J.; Kerkhof, E.; et al. Multi-hallmark long noncoding RNA maps reveal non-small cell lung cancer vulnerabilities. Cell Genom. 2022, 2, 100171.
- Pacholewska, A.; Sung, M.H. lncRNA expression predicts mRNA abundance. Epigenomics 2019, 11, 1121–1128.
- Yang, M.; Lu, H.; Liu, J.; Wu, S.; Kim, P.; Zhou, X. lncRNAfunc: A knowledgebase of lncRNA function in human cancer. Nucleic Acids Res. 2022, 50, D1295–D1306.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
671
Revisions:
2 times
(View History)
Update Date:
25 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




