
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Diletta Fontana | -- | 3842 | 2023-06-20 14:35:44 | | | |
| 2 | Jessie Wu | + 12 word(s) | 3854 | 2023-06-21 03:01:42 | | |
Video Upload Options
The myelodysplastic syndromes/myeloproliferative overlap neoplasms (MDS/MPN) comprise a heterogeneous group of myeloid neoplastic diseases with clinical and pathologic overlapping features of both myelodysplastic and myeloproliferative neoplasms. The 2016 World Health Organization (WHO) classification included five entities: chronic myelomonocytic leukemia (CMML), atypical CML BCR-ABL1− (aCML), juvenile myelomonocytic leukemia (JMML), MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T), and MDS/MPN unclassifiable (MDS/MPN-U). In 2022, there emerged two competing classifications for myeloid neoplasms: the International Consensus Classification (ICC) and the fifth edition of the WHO classification. Both classifications now expand on these categories; in particular, MDS/MPN with ring sideroblasts and thrombocytosis (MDS/MPN-RS-T) has been split into two entities in the ICC 2022 based on the presence/absence of the SF3B1 mutation. Moreover, both the WHO 2022 classification and the ICC move JMML to be grouped with pediatric and/or germline mutation-associated disorders.
1. Myelodysplastic Syndromes/Myeloproliferative Neoplasms with Neutrophilia (Also Known as Atypical CML)
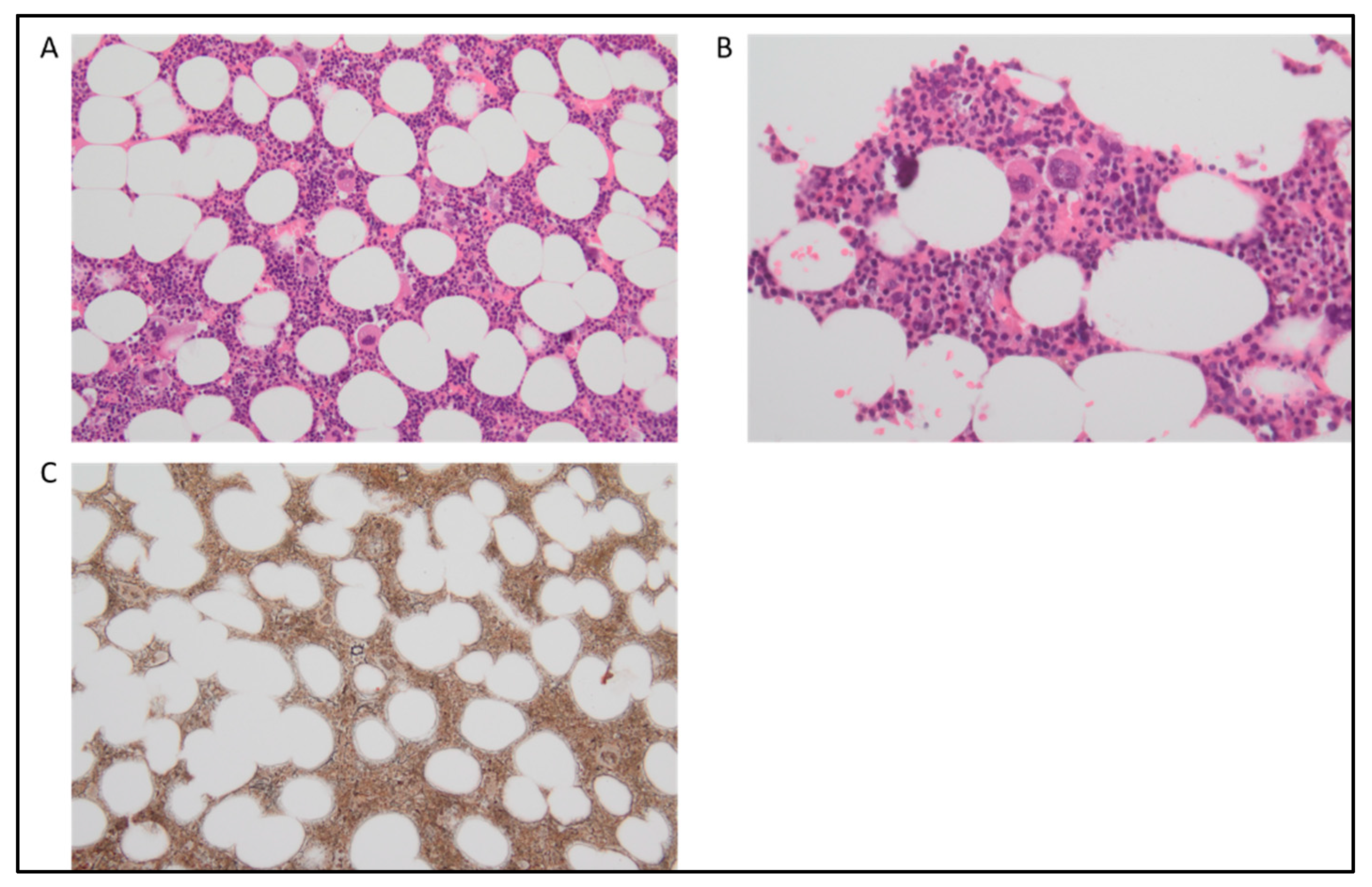
2. Myelodysplastic Syndromes/Myeloproliferative Overlap Neoplasms with SF3B1 Mutation and Thrombocytosis and MDS/MPN with Ring Sideroblasts and Thrombocytosis
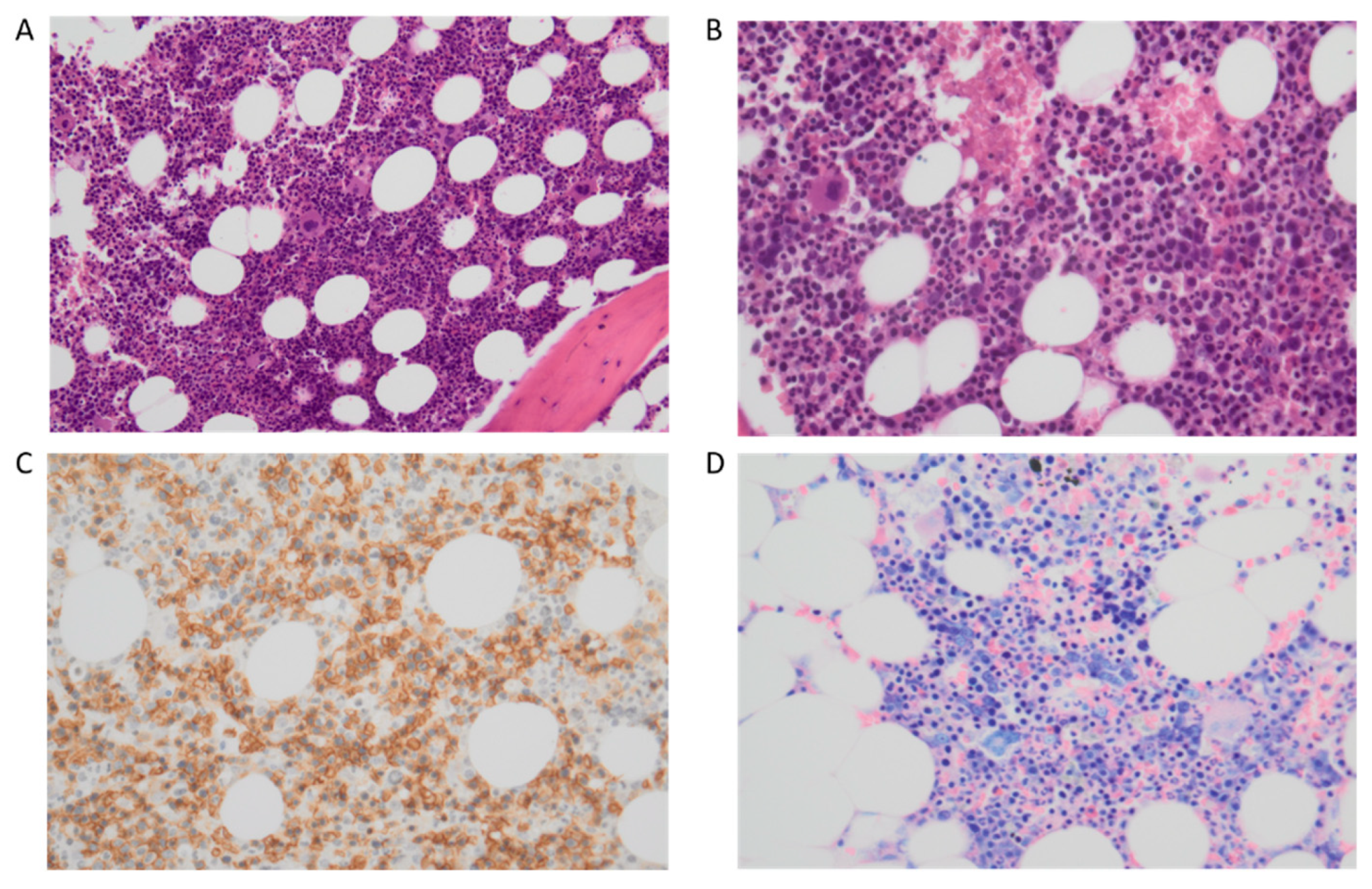
3. Myelodysplastic/Myeloproliferative Neoplasm, Not Otherwise Specified
4. Chronic Myelomonocytic Leukemia
4.1. Cytogenetics and Molecular Genetics
4.2. Flow Cytometry Immunophenotyping
4.3. Diagnosis and Differential Diagnosis of Chronic Myelomonocytic Leukemia
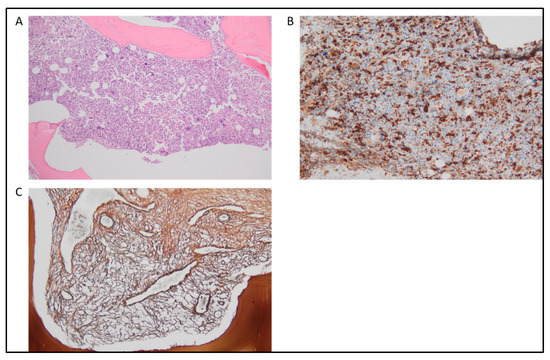
5. Clonal Monocytosis of Undetermined Significance and Clonal Cytopenia with Monocytosis of Undetermined Significance
References
- Khoury, J.D.; Solary, E.; Abla, O.; Akkari, Y.; Alaggio, R.; Apperley, J.F.; Bejar, R.; Berti, E.; Busque, L.; Chan, J.K.C.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Myeloid and Histiocytic/Dendritic Neoplasms. Leukemia 2022, 36, 1703–1719.
- Arber, D.A.; Orazi, A.; Hasserjian, R.P.; Borowitz, M.J.; Calvo, K.R.; Kvasnicka, H.M.; Wang, S.A.; Bagg, A.; Barbui, T.; Branford, S.; et al. International Consensus Classification of Myeloid Neoplasms and Acute Leukemias: Integrating morphologic, clinical, and genomic data. Blood 2022, 140, 1200–1228.
- Patnaik, M.M.; Barraco, D.; Lasho, T.L.; Finke, C.M.; Reichard, K.; Hoversten, K.P.; Ketterling, R.P.; Gangat, N.; Tefferi, A. Targeted next generation sequencing and identification of risk factors in World Health Organization defined atypical chronic myeloid leukemia. Am. J. Hematol. 2017, 92, 542–548.
- Vardiman, J.W.; Bain, B.; Inbert, M.; Brunning, R.D.; Pierre, R.V.; Flandrin, G. Atypical chronic myeloid leukemia. In WHO Classification of Tumours: Pathology and Genetics of Tumours of Haematopoietic and Lymphoid Tissues; Jaffe, E., Harris, N.L., Stein, H., Vardiman, J., Eds.; IARC Press: Lyon, France, 2001; pp. 53–57.
- Breccia, M.; Biondo, F.; Latagliata, R.; Carmosino, I.; Mandelli, F.; Alimena, G. Identification of risk factors in atypical chronic myeloid leukemia. Haematologica 2006, 91, 1566–1568.
- Oscier, D. Atypical chronic myeloid leukemias. Pathol. Biol. 1997, 45, 587–593.
- Cazzola, M.; Malcovati, L.; Invernizzi, R. Myelodysplastic/myeloproliferative neoplasms. Hematol. Am. Soc. Hematol. Educ. Program 2011, 1, 264–272.
- Hernandez, J.; Villaescusa, T.; Arefi, M.; López, L.; Garcia, J. Atypical Chronic Myeloid Leukemia (aCML). Atlas Genet. Cytogenet. Oncol. Haematol. 2009, 13, 432–433.
- Zhang, H.; Wilmot, B.; Bottomly, D.; Dao, K.T.; Stevens, E.; Eide, C.A.; Khanna, V.; Rofelty, A.; Savage, S.; Reister Schultz, A.; et al. Genomic landscape of neutrophilic leukemias of ambiguous diagnosis. Blood 2019, 134, 867–879.
- Fontana, D.; Ramazzotti, D.; Aroldi, A.; Redaelli, S.; Magistroni, V.; Pirola, A.; Niro, A.; Massimino, L.; Mastini, C.; Brambilla, V.; et al. Integrated Genomic, Functional, and Prognostic Characterization of Atypical Chronic Myeloid Leukemia. Hemasphere 2020, 4, e497.
- Giri, S.; Pathak, R.; Martin, M.G.; Bhatt, V.R. Characteristics and survival of BCR/ABL negative chronic myeloid leukemia: A retrospective analysis of the Surveillance, Epidemiology and End Results database. Ther. Adv. Hematol. 2015, 6, 308–312.
- Orazi, A.; Bennet, J.M.; Bain, B.J.; Brunning, R.D.; Thiele, J. Atypical chronic myeloid leukaemia. In WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues, 4th ed.; Swerdlow, S.H., Campo, E., Harris, N.L., Jaffe, E.S., Pileri, S.A., Stein, H., Thiele, J., Eds.; IARC Press: Lyon, France, 2017; pp. 87–89.
- Vardiman, J.W.; Thiele, J.; Arber, D.A.; Brunning, R.D.; Borowitz, M.J.; Porwit, A.; Harris, N.L.; Le Beau, M.M.; Hellström-Lindberg, E.; Tefferi, A.; et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: Rationale and important changes. Blood 2009, 114, 937–951.
- Crisà, E.; Nicolosi, M.; Ferri, V.; Favini, C.; Gaidano, G.; Patriarca, A. Atypical Chronic Myeloid Leukemia: Where Are We Now? Int. J. Mol. Sci. 2020, 21, 6862.
- Fontana, D.; Gambacorti-Passerini, C.; Piazza, R. Molecular Pathogenesis of BCR-ABL-Negative Atypical Chronic Myeloid Leukemia. Front. Oncol. 2021, 11, 756348.
- Arber, D.A.; Orazi, A.; Hasserjian, R.; Thiele, J.; Borowitz, M.J.; Le Beau, M.M.; Bloomfield, C.D.; Cazzola, M.; Vardiman, J.W. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood 2016, 127, 2391–2405.
- Kurzrock, R.; Bueso-Ramos, C.E.; Kantarjian, H.; Freireich, E.; Tucker, S.L.; Siciliano, M.; Pilat, S.; Talpaz, M. BCR rearrangement-negative chronic myelogenous leukemia revisited. J. Clin. Oncol. 2001, 19, 2915–2926.
- Patnaik, M.M.; Tefferi, A. Atypical chronic myeloid leukemia and myelodysplastic/myeloproliferative neoplasm, not otherwise specified: 2023 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2023, 98, 681–689.
- Gotlib, J.; Maxson, J.E.; George, T.I.; Tyner, J.W. The new genetics of chronic neutrophilic leukemia and atypical CML: Implications for diagnosis and treatment. Blood 2013, 122, 1707–1711.
- Orazi, A.; Germing, U. The myelodysplastic/myeloproliferative neoplasms: Myeloproliferative diseases with dysplastic features. Leukemia 2008, 22, 1308–1319.
- Maxson, J.E.; Tyner, J.W. Genomics of chronic neutrophilic leukemia. Blood 2017, 129, 715–722.
- Dao, K.H.; Tyner, J.W. What’s different about atypical CML and chronic neutrophilic leukemia? Hematol. Am. Soc. Hematol. Educ. Program 2015, 2015, 264–271.
- Maxson, J.E.; Gotlib, J.; Pollyea, D.A.; Fleischman, A.G.; Agarwal, A.; Eide, C.A.; Bottomly, D.; Wilmot, B.; McWeeney, S.K.; Tognon, C.E.; et al. Oncogenic CSF3R Mutations in Chronic Neutrophilic Leukemia and Atypical CML. N. Engl. J. Med. 2013, 368, 1781–1790.
- Schwartz, L.C.; Mascarenhas, J. Current and evolving understanding of atypical chronic myeloid leukemia. Blood Rev. 2019, 33, 74–81.
- Carreño-Tarragona, G.; Alvarez-Larran, A.; Harrison, C.N.; Martínez-Ávila, J.C.; Hernandez-Boluda, J.C.; Ferrer-Marin, F.; Radia, D.H.; Mora Casterá, E.; Francis, S.; González-Martínez, T.; et al. CNL and aCML should be considered as single entity based on molecular profiles and outcomes. Blood Adv. 2022, 9, 1672–1681.
- Thomopoulos, T.P.; Symeonidis, A.; Kourakli, A.; Papageorgiou, S.G.; Pappa, V. Chronic Neutrophilic Leukemia: A Comprehensive Review of Clinical Characteristics, Genetic Landscape and Management. Front. Oncol. 2022, 12, 891961.
- Thakral, B.; Anastasi, J.; Wang, S.A. 17-Myeloproliferative and “Overlap” Myelodysplastic/Myeloproliferative Neoplasms. Found. Diagn. Pathol. 2018, 488–538.e484.
- Wang, S.A.; Hasserjian, R.P.; Fox, P.S.; Rogers, H.J.; Geyer, J.T.; Chabot-Richards, D.; Weinzierl, E.; Hatem, J.; Jaso, J.; Kanagal-Shamanna, R.; et al. Atypical chronic myeloid leukemia is clinically distinct from unclassifiable myelodysplastic/myeloproliferative neoplasms. Blood 2014, 123, 2645–2651.
- Wang, S.A.; Hasserjian, R.P.; Tam, W.; Tsai, A.G.; Geyer, J.T.; George, T.I.; Foucar, K.; Rogers, H.J.; Hsi, E.D.; Rea, B.A.; et al. Bone marrow morphology is a strong discriminator between chronic eosinophilic leukemia, not otherwise specified and reactive idiopathic hypereosinophilic syndrome. Haematologica 2017, 102, 1352–1360.
- Fend, F.; Horn, T.; Koch, I.; Vela, T.; Orazi, A. Atypical chronic myeloid leukemia as defined in the WHO classification is a JAK2 V617F negative neoplasm. Leuk. Res. 2008, 32, 1931–1935.
- Gambacorti-Passerini, C.B.; Donadoni, C.; Parmiani, A.; Pirola, A.; Redaelli, S.; Signore, G.; Piazza, V.; Malcovati, L.; Fontana, D.; Spinelli, R.; et al. Recurrent ETNK1 mutations in atypical chronic myeloid leukemia. Blood 2015, 125, 499–503.
- Fontana, D.; Mauri, M.; Renso, R.; Docci, M.; Crespiatico, I.; Røst, L.M.; Jang, M.; Niro, A.; D’Aliberti, D.; Massimino, L.; et al. ETNK1 mutations induce a mutator phenotype that can be reverted with phosphoethanolamine. Nat. Commun. 2020, 11, 5938.
- Fontana, D.; Gambacorti-Passerini, C.; Piazza, R. Impact of ETNK1 somatic mutations on phosphoethanolamine synthesis, ROS production and DNA damage. Mol. Cell. Oncol. 2021, 8, 1877598.
- Piazza, R.; Valletta, S.; Winkelmann, N.; Redaelli, S.; Spinelli, R.; Pirola, A.; Antolini, L.; Mologni, L.; Donadoni, C.; Papaemmanuil, E.; et al. Recurrent SETBP1 mutations in atypical chronic myeloid leukemia. Nat. Genet. 2013, 45, 18–24.
- Piazza, R.; Magistroni, V.; Redaelli, S.; Mauri, M.; Massimino, L.; Sessa, A.; Peronaci, M.; Lalowski, M.; Soliymani, R.; Mezzatesta, C.; et al. SETBP1 induces transcription of a network of development genes by acting as an epigenetic hub. Nat. Commun. 2018, 9, 2192.
- Meggendorfer, M.; Bacher, U.; Alpermann, T.; Haferlach, C.; Kern, W.; Gambacorti-Passerini, C.; Haferlach, T.; Schnittger, S. SETBP1 mutations occur in 9% of MDS/MPN and in 4% of MPN cases and are strongly associated with atypical CML, monosomy 7, isochromosome i(17)(q10), ASXL1 and CBL mutations. Leukemia 2013, 27, 1852–1860.
- Dhakal, P.; Gundabolu, K.; Amador, C.; Rayamajhi, S.; Bhatt, V.R. Atypical chronic myeloid leukemia: A rare entity with management challenges. Future Oncol. 2018, 14, 177–185.
- Gotlib, J. How I treat atypical chronic myeloid leukemia. Blood 2017, 129, 838–845.
- Carulli, G.; Fabiani, O.; Azzara, A. The syndrome of abnormal chromatin clumping in leukocytes. Haematologica 1997, 82, 635–636.
- Tefferi, A.; Elliot, M.A.; Pardanani, A. Atypical myeloproliferative disorders: Diagnosis and management. Mayo Clin. Proc. 2006, 81, 553–563.
- Jones, A.V.; Kreil, S.; Zoi, K.; Waghorn, K.; Curtis, C.; Zhang, L.Y.; Score, J.; Seear, R.; Chase, A.J.; Grand, F.H.; et al. Widespread occurrence of the JAK2 V617F mutation in chronic myeloproliferative disorders. Blood 2005, 106, 2162–2168.
- Mughal, T.I.; Cross, N.C.; Padron, E.; Tiu, R.V.; Savona, M.; Malcovati, L.; Tibes, R.; Komrokji, R.S.; Kiladjian, J.J.; Garcia-Manero, G.; et al. An International MDS/MPN Working Group’s perspective and recommendations on molecular pathogenesis, diagnosis and clinical characterization of myelodysplastic/myeloproliferative neoplasms. Haematologica 2015, 100, 1117–1130.
- Tiu, R.V.; Sekeres, M.A. Making sense of the myelodysplastic/myeloproliferative neoplasms overlap syndromes. Curr. Opin. Hematol. 2014, 21, 131–140.
- Zoi, K.; Cross, N.C. Molecular pathogenesis of atypical CML, CMML and MDS/MPN-unclassifiable. Int. J. Hematol. 2015, 101, 229–242.
- Meggendorfer, M.; Haferlach, T.; Alpermann, T.; Jeromin, S.; Haferlach, C.; Kern, W.; Schnittger, S. Specific molecular mutation patterns delineate chronic neutrophilic leukemia, atypical chronic myeloid leukemia, and chronic myelomonocytic leukemia. Haematologica 2014, 99, e244–e246.
- Makishima, H.; Yoshida, K.; Nguyen, N.; Przychodzen, B.; Sanada, M.; Okuno, Y.; Ng, K.P.; Gudmundsson, K.O.; Vishwakarma, B.A.; Jerez, A.; et al. Somatic SETBP1 mutations in myeloid malignancies. Nat. Genet. 2013, 45, 942–946.
- Abdel-Wahab, O.; Mullally, A.; Hedvat, C.; Garcia-Manero, G.; Patel, J.; Wadleigh, M.; Malinge, S.; Yao, J.; Kilpivaara, O.; Bhat, R.; et al. Genetic characterization of TET1, TET2, and TET3 alterations in myeloid malignancies. Blood 2009, 114, 144–147.
- Patnaik, M.M.; Lasho, T. Myelodysplastic syndrome/myeloproliferative neoplasm overlap syndromes: A focused review. Hematol. Am. Soc. Hematol. Educ. Program 2020, 2020, 460–464.
- Elena, C.; Gallì, A.; Such, E.; Meggendorfer, M.; Germing, U.; Rizzo, E.; Cervera, J.; Molteni, E.; Fasan, A.; Schuler, E.; et al. Integrating clinical features and genetic lesions in the risk assessment of patients with chronic myelomonocytic leukemia. Blood 2016, 128, 1408–1417.
- Itzykson, R.; Fenaux, P.; Bowen, D.; Cross, N.C.P.; Cortes, J.; De Witte, T.; Germing, U.; Onida, F.; Padron, E.; Platzbecker, U.; et al. Diagnosis and Treatment of Chronic Myelomonocytic Leukemias in Adults: Recommendations From the European Hematology Association and the European LeukemiaNet. Hemasphere 2018, 2, e150.
- Mason, C.C.; Khorashad, J.S.; Tantravahi, S.K.; Kelley, T.W.; Zabriskie, M.S.; Yan, D.; Pomicter, A.D.; Reynolds, K.R.; Eiring, A.M.; Kronenberg, Z.; et al. Age-related mutations and chronic myelomonocytic leukemia. Leukemia 2016, 30, 906–913.
- Palomo, L.; Ibáñez, M.; Abáigar, M.; Vázquez, I.; Álvarez, S.; Cabezón, M.; Tazón-Vega, B.; Rapado, I.; Fuster-Tormo, F.; Cervera, J.; et al. Spanish Guidelines for the use of targeted deep sequencing in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br. J. Haematol. 2020, 188, 605–622.
- Woo, J.; Choi, D.R.; Storer, B.E.; Yeung, C.; Halpern, A.B.; Salit, R.B.; Sorror, M.L.; Woolston, D.W.; Monahan, T.; Scott, B.L.; et al. Impact of clinical, cytogenetic, and molecular profiles on long-term survival after transplantation in patients with chronic myelomonocytic leukemia. Haematologica 2020, 105, 652–660.
- Merlevede, J.; Droin, N.; Qin, T.; Meldi, K.; Yoshida, K.; Morabito, M.; Chautard, E.; Auboeuf, D.; Fenaux, P.; Braun, T.; et al. Mutation allele burden remains unchanged in chronic myelomonocytic leukaemia responding to hypomethylating agents. Nat. Commun. 2016, 7, 10767.
- Cargo, C.; Cullen, M.; Taylor, J.; Short, M.; Glover, P.; Van Hoppe, S.; Smith, A.; Evans, P.; Crouch, S. The use of targeted sequencing and flow cytometry to identify patients with a clinically significant monocytosis. Blood 2019, 133, 1325–1334.
- Palomo, L.; Meggendorfer, M.; Hutter, S.; Twardziok, S.; Ademà, V.; Fuhrmann, I.; Fuster-Tormo, F.; Xicoy, B.; Zamora, L.; Acha, P.; et al. Molecular landscape and clonal architecture of adult myelodysplastic/myeloproliferative neoplasms. Blood 2020, 136, 1851–1862.
- Vallapureddy, R.; Lasho, T.L.; Hoversten, K.; Finke, C.M.; Ketterling, R.; Hanson, C.; Gangat, N.; Tefferi, A.; Patnaik, M.M. Nucleophosmin 1 (NPM1) mutations in chronic myelomonocytic leukemia and their prognostic relevance. Am. J. Hematol. 2017, 92, E614–E618.
- Ball, M.; List, A.F.; Padron, E. When clinical heterogeneity exceeds genetic heterogeneity: Thinking outside the genomic box in chronic myelomonocytic leukemia. Blood 2016, 128, 2381–2387.
- Meggendorfer, M.; Roller, A.; Haferlach, T.; Eder, C.; Dicker, F.; Grossmann, V.; Kohlmann, A.; Alpermann, T.; Yoshida, K.; Ogawa, S.; et al. SRSF2 mutations in 275 cases with chronic myelomonocytic leukemia (CMML). Blood 2012, 120, 3080–3088.
- Itzykson, R.; Kosmider, O.; Renneville, A.; Gelsi-Boyer, V.; Meggendorfer, M.; Morabito, M.; Berthon, C.; Adès, L.; Fenaux, P.; Beyne-Rauzy, O.; et al. Prognostic score including gene mutations in chronic myelomonocytic leukemia. J. Clin. Oncol. 2013, 31, 2428–2436.
- Malcovati, L.; Papaemmanuil, E.; Ambaglio, I.; Elena, C.; Gallì, A.; Della Porta, M.G.; Travaglino, E.; Pietra, D.; Pascutto, C.; Ubezio, M.; et al. Driver somatic mutations identify distinct disease entities within myeloid neoplasms with myelodysplasia. Blood 2014, 124, 1513–1521.
- Ricci, C.; Fermo, E.; Corti, S.; Molteni, M.; Faricciotti, A.; Cortelezzi, A.; Lambertenghi Deliliers, G.; Beran, M.; Onida, F. RAS mutations contribute to evolution of chronic myelomonocytic leukemia to the proliferative variant. Clin. Cancer Res. 2010, 16, 2246–2256.
- Kohlmann, A.; Grossmann, V.; Klein, H.U.; Schindela, S.; Weiss, T.; Kazak, B.; Dicker, F.; Schnittger, S.; Dugas, M.; Kern, W.; et al. Next-generation sequencing technology reveals a characteristic pattern of molecular mutations in 72.8% of chronic myelomonocytic leukemia by detecting frequent alterations in TET2, CBL, RAS, and RUNX1. J. Clin. Oncol. 2010, 28, 3858–3865.
- Patnaik, M.M.; Lasho, T.L.; Vijayvargiya, P.; Finke, C.M.; Hanson, C.A.; Ketterling, R.P.; Gangat, N.; Tefferi, A. Prognostic interaction between ASXL1 and TET2 mutations in chronic myelomonocytic leukemia. Blood Cancer J. 2016, 6, e385.
- Steensma, D.P.; Dewald, G.W.; Lasho, T.L.; Powell, H.L.; McClure, R.F.; Levine, R.L.; Gilliland, D.G.; Tefferi, A. The JAK2 V617F activating tyrosine kinase mutation is an infrequent event in both “atypical” myeloproliferative disorders and myelodysplastic syndromes. Blood 2005, 106, 1207–1209.
- Itzykson, R.; Kosmider, O.; Renneville, A.; Morabito, M.; Preudhomme, C.; Berthon, C.; Adès, L.; Fenaux, P.; Platzbecker, U.; Gagey, O.; et al. Clonal architecture of chronic myelomonocytic leukemias. Blood 2013, 121, 2186–2198.
- Beran, M.; Shen, Y.; Onida, F.; Wen, S.; Kantarjian, H.; Estey, E. Prognostic significance of monocytosis in patients with myeloproliferative disorders. Leuk Lymphoma 2006, 47, 417–423.
- Meggendorfer, M.; Jeromin, S.; Haferlach, C.; Kern, W.; Haferlach, T. The mutational landscape of 18 investigated genes clearly separates four subtypes of myelodysplastic/myeloproliferative neoplasms. Haematologica 2018, 103, e192–e195.
- Peng, J.; Zuo, Z.; Fu, B.; Oki, Y.; Tang, G.; Goswami, M.; Priyanka, P.; Muzzafar, T.; Medeiros, L.J.; Luthra, R.; et al. Chronic myelomonocytic leukemia with nucleophosmin (NPM1) mutation. Eur. J. Haematol. 2016, 96, 65–71.
- Ernst, T.; Chase, A.; Zoi, K.; Waghorn, K.; Hidalgo-Curtis, C.; Score, J.; Jones, A.; Grand, F.; Reiter, A.; Hochhaus, A.; et al. Transcription factor mutations in myelodysplastic/myeloproliferative neoplasms. Haematologica 2010, 95, 1473–1480.
- Ziegler-Heitbrock, L.; Ancuta, P.; Crowe, S.; Dalod, M.; Grau, V.; Hart, D.N.; Leenen, P.J.; Liu, Y.J.; MacPherson, G.; Randolph, G.J.; et al. Nomenclature of monocytes and dendritic cells in blood. Blood 2010, 116, e74–e80.
- Selimoglu-Buet, D.; Wagner-Ballon, O.; Saada, V.; Bardet, V.; Itzykson, R.; Bencheikh, L.; Morabito, M.; Met, E.; Debord, C.; Benayoun, E.; et al. Characteristic repartition of monocyte subsets as a diagnostic signature of chronic myelomonocytic leukemia. Blood 2015, 125, 3618–3626.
- Barraco, D.; Cerquozzi, S.; Gangat, N.; Patnaik, M.M.; Lasho, T.; Finke, C.; Hanson, C.A.; Ketterling, R.P.; Pardanani, A.; Tefferi, A. Monocytosis in polycythemia vera: Clinical and molecular correlates. Am. J. Hematol. 2017, 92, 640–645.
- Tefferi, A.; Shah, S.; Mudireddy, M.; Lasho, T.L.; Barraco, D.; Hanson, C.A.; Ketterling, R.P.; Elliott, M.A.; Patnaik, M.S.; Pardanani, A.; et al. Monocytosis is a powerful and independent predictor of inferior survival in primary myelofibrosis. Br. J. Haematol. 2018, 183, 835–838.
- Patnaik, M.M. How I diagnose and treat chronic myelomonocytic leukemia. Haematologica 2022, 107, 1503–1517.
- Wan, Z.; Han, B. Comparison and Implications of Mutational Profiles of Myelodysplastic Syndromes, Myeloproliferative Neoplasms, and Myelodysplastic/Myeloproliferative Neoplasms: A Meta-Analysis. Front. Oncol. 2020, 10, 579221.
- Valent, P.; Orazi, A.; Savona, M.R.; Patnaik, M.M.; Onida, F.; van de Loosdrecht, A.A.; Haase, D.; Haferlach, T.; Elena, C.; Pleyer, L.; et al. Proposed diagnostic criteria for classical chronic myelomonocytic leukemia (CMML), CMML variants and pre-CMML conditions. Haematologica 2019, 104, 1935–1949.




