
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Muhammad Adnan | -- | 2710 | 2023-06-15 15:15:07 | | | |
| 2 | Wendy Huang | Meta information modification | 2710 | 2023-06-16 09:34:34 | | |
Video Upload Options
Fungi are an important group of microorganisms that play crucial roles in a variety of ecological and biotechnological processes. Fungi depend on intracellular protein trafficking, which involves moving proteins from their site of synthesis to the final destination within or outside the cell. The soluble N-ethylmaleimide-sensitive factor attachment protein receptors (SNARE) proteins are vital components of vesicle trafficking and membrane fusion, ultimately leading to the release of cargos to the target destination. The v-SNARE (vesicle-associated SNARE) Snc1 is responsible for anterograde and retrograde vesicle trafficking between the plasma membrane (PM) and Golgi. It allows for the fusion of exocytic vesicles to the PM and the subsequent recycling of Golgi-localized proteins back to the Golgi via three distinct and parallel recycling pathways. This recycling process requires several components, including a phospholipid flippase (Drs2-Cdc50), an F-box protein (Rcy1), a sorting nexin (Snx4-Atg20), a retromer submit, and the COPI coat complex. Snc1 interacts with exocytic SNAREs (Sso1/2, Sec9) and the exocytic complex to complete the process of exocytosis. It also interacts with endocytic SNAREs (Tlg1 and Tlg2) during endocytic trafficking. Snc1 has been extensively investigated in fungi and has been found to play crucial roles in various aspects of intracellular protein trafficking.
1. Introduction
| Protein Name | Function | Interaction with SNC1 | References |
|---|---|---|---|
| Sso1/Sso2 | Vesicle fusion with plasma membrane | Essential for exocytosis of secretory vesicles | [20] |
| Sec1/Munc18 | Docking of secretory vesicles and their fusion with the PM | Critical for efficient vesicle fusion with plasma membrane | [12] |
| Sec9 | Docking of cargo vesicles and their fusion | Fusion of the secretory vesicles and PM for efficient exocytosis | [20][21] |
| Sec18/NSF Sec17/SNAP |
Assembly and disassembly of SNAREs | Assembly and disassembly of SNC1 mediated SNAREs | [22] |
| Exo70/ Exo84 |
Important components of exocyst complex | Promotes SNC1 localization towards the exocytic sites on the PM | [23][24] |
| Sec3 | Important components of the exocyst complex | Promote exocytic complex formation at the PM | [23][24] |
| Sec5 | |||
| Sec6 | |||
| Sec8 | |||
| Sec15 | |||
| Sro7/Sro77 | Exocytosis and actin organization | Regulate trafficking and localization of SNC1 to specific membrane domains | [25] |
| Cdc42 | Subunit of exocyst complex | Fusion of secretory vesicles and the PM | [10] |
| Drs2-Cdc50 | Phospholipid flippase (Subunit of exocyst complex) | Post-endocytic recycling of SNC1 | [16][26] |
| Rcy1 | F-box protein (Subunit of exocyst complex) | ||
| Snx4-Atg20 | sorting nexin (Subunit of exocyst complex) | ||
| COPI coat complex | COPI coat complex surrounds the cargo vesicles for cellular transportation | ||
| ArfA | Recruitment of COPI to Golgi membranes | Regulate SNC1 localization and activity in the secretory pathway | [27] |
| AP180 | Functional role in endocytosis | Play a cargo-specific role in SNC1 internalization | [28] |
| Tlg1/Tlg2 | t-SNAREs localized to late Golgi and endosomes | Recycling of Snc1 protein from the PM towards Golgi | [22] |
| YPT1 | GTPase involved in vesicle trafficking | Facilitates the fusion of vesicles | [12] |
| Sla1 | Endocytosis and actin organization | SNC1 and Sla1 interaction regulates the internalization of the α-factor receptor Ste2 | [29] |
| End3/End4 | Endocytosis and actin organization | Efficient endocytosis of a subset of membrane proteins | [30] |
| Vam3/Vam7 | Vesicle fusion | Efficient fusion of vesicles with the vacuole | [22][31] |
| Pep12 | Endosomal and vacuolar trafficking | Essential for Snc1 recycling from endosomes to PM | [26][32] |
| Vps41 | Key regulator of SNC1/2 recycling | Interacts with SNC1/2 and promotes its sorting into recycling vesicles | [33] |
| Ede1 | Endocytic proteins | Mediate vesicle fusion during endocytosis | [34][35] |
| Syp1 | |||
| Pal1 | |||
| Vti1 | Trans-Golgi network to endosome transportation of vesicles | Fusion of secretory vesicles and the PM | [26][32] |
| Sec22 | ER-to-Golgi vesicular transport | Can complement SNC1 in vivo | [36][37] |
| Nyv1 | Fusion of vesicles with the vacuole | Efficient vesicle fusion with the vacuole | [22][32] |
| Syn8 | Endosomal t-SNARE involved in vesicle fusion | Exhibits promiscuous interactions with Snc1/2 | [32] |
| Sed5 | t-SNAREs of cis-Golgi | Mediate fusion of initial PM derived vesicles | [38] |
| RSP5 | Ubiquitin ligase | Efficient recycling of SNC1 | [30] |
| Rab1 | Rab-GTPase Regulate vesicular trafficking | Rab1 inactivation blocks SNC1 recycling from Golgi to PM | [39] |
| Rab7 | Rab-GTPase Regulate vesicular trafficking | Regulate SNC1 trafficking from trans-Golgi network to PM and vice versa | [40] |
| Ras2 | Ras protein required to activate adenylate cyclase pathway | Ras2-SNC1 interaction suggest a role in response to nutrient availability | [41] |
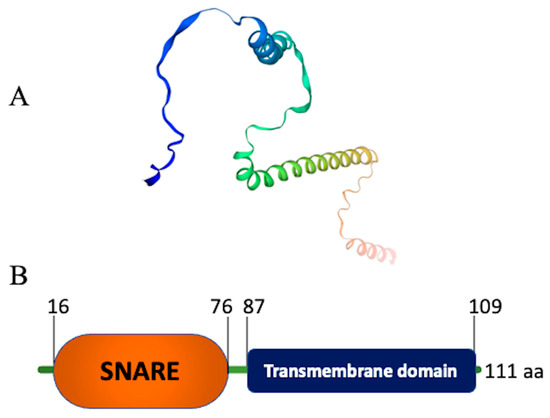
2. Structure of Snc1 Protein
3. Exocytosis
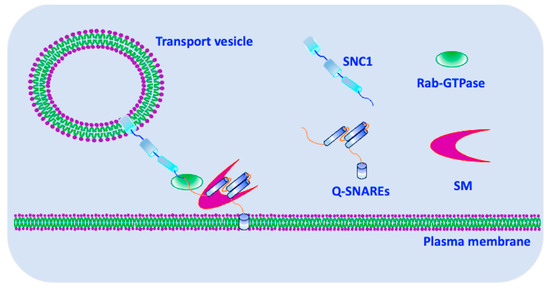
4. Endocytosis
References
- Pimentel, P.S.S.-R.; de Oliveira, J.B.; Astolfi-Filho, S.; Pereira, N., Jr. Enzymatic Hydrolysis of Lignocellulosic Biomass Using an Optimized Enzymatic Cocktail Prepared from Secretomes of Filamentous Fungi Isolated from Amazonian Biodiversity. Appl. Biochem. Biotechnol. 2021, 193, 3915–3935.
- Wang, Y.; Zheng, X.; Li, G.; Wang, X. TORC1 Signaling in Fungi: From Yeasts to Filamentous Fungi. Microorganisms 2023, 11, 218.
- Staples, M.I.; Frazer, C.; Fawzi, N.L.; Bennett, R.J. Phase Separation in Fungi. Nat. Microbiol. 2023, 8, 375–386.
- Fitz, E.; Wanka, F.; Seiboth, B. The Promoter Toolbox for Recombinant Gene Expression in Trichoderma Reesei. Front. Bioeng. Biotechnol. 2018, 6, 135.
- Choi, U.B.; Dunleavy, K.; Matlock, H.; Gething, C.; Howells, G.; Misra, B.; White, K.I.; Brunger, A. Conformational Dynamics of SNARE Recycling Mediated by NSF. FASEB J. 2022, 36.
- Adnan, M.; Islam, W.; Zhang, J.; Zheng, W.; Lu, G.-D. Diverse Role of SNARE Protein Sec22 in Vesicle Trafficking, Membrane Fusion, and Autophagy. Cells 2019, 8, 337.
- Adnan, M.; Fang, W.; Sun, P.; Zheng, Y.; Abubakar, Y.S.; Zhang, J.; Lou, Y.; Zheng, W.; Lu, G. R-SNARE FgSec22 Is Essential for Growth, Pathogenicity and DON Production of Fusarium Graminearum. Curr. Genet. 2020, 66, 421–435.
- Adnan, M.; Islam, W.; Noman, A.; Hussain, A.; Anwar, M.; Khan, M.U.; Akram, W.; Ashraf, M.F.; Raza, M.F. Q-SNARE Protein FgSyn8 Plays Important Role in Growth, DON Production and Pathogenicity of Fusarium Graminearum. Microb. Pathog. 2020, 140, 103948.
- Valkonen, M.; Kalkman, E.R.; Saloheimo, M.; Penttilaö, M.; Read, N.D.; Duncan, R.R. Spatially Segregated SNARE Protein Interactions in Living Fungal Cells. J. Biol. Chem. 2007, 282, 22775–22785.
- He, B.; Guo, W. The Exocyst Complex in Polarized Exocytosis. Curr. Opin. Cell Biol. 2009, 21, 537–542.
- Aalto, M.K.; Ronne, H.; Keränen, S. Yeast Syntaxins Sso1p and Sso2p Belong to a Family of Related Membrane Proteins That Function in Vesicular Transport. EMBO J. 1993, 12, 4095–4104.
- Hong, W.; Lev, S. Tethering the Assembly of SNARE Complexes. Trends Cell Biol. 2014, 24, 35–43.
- Rizo, J.; Sari, L.; Qi, Y.; Im, W.; Lin, M.M. All-Atom Molecular Dynamics Simulations of Synaptotagmin-SNARE-Complexin Complexes Bridging a Vesicle and a Flat Lipid Bilayer. Elife 2022, 11, e76356.
- Wang, S.; Ma, C. Neuronal SNARE Complex Assembly Guided by Munc18-1 and Munc13-1. FEBS Open Bio 2022, 12, 1939–1957.
- Fischer, R.; Zekert, N.; Takeshita, N. Polarized Growth in Fungi–Interplay between the Cytoskeleton, Positional Markers and Membrane Domains. Mol. Microbiol. 2008, 68, 813–826.
- Best, J.T.; Xu, P.; McGuire, J.G.; Leahy, S.N.; Graham, T.R. Yeast Synaptobrevin, Snc1, Engages Distinct Routes of Postendocytic Recycling Mediated by a Sorting Nexin, Rcy1-COPI, and Retromer. Mol. Biol. Cell 2020, 31, 944–962.
- Fiedler, M.R.; Barthel, L.; Kubisch, C.; Nai, C.; Meyer, V. Construction of an Improved Aspergillus Niger Platform for Enhanced Glucoamylase Secretion. Microb. Cell Factories 2018, 17, 1–12.
- Wu, Y.; Sun, X.; Xue, X.; Luo, H.; Yao, B.; Xie, X.; Su, X. Overexpressing Key Component Genes of the Secretion Pathway for Enhanced Secretion of an Aspergillus Niger Glucose Oxidase in Trichoderma Reesei. Enzym. Microb. Technol. 2017, 106, 83–87.
- Yang, S.; Zhou, X.; Guo, P.; Lin, Y.; Fan, Q.; Zuriegat, Q.; Lu, S.; Yang, J.; Yu, W.; Liu, H. The Exocyst Regulates Hydrolytic Enzyme Secretion at Hyphal Tips and Septa in the Banana Fusarium Wilt Fungus Fusarium Odoratissimum. Appl. Environ. Microbiol. 2021, 87, e03088-20.
- Kubicek, C.P.; Starr, T.L.; Glass, N.L. Plant Cell Wall–Degrading Enzymes and Their Secretion in Plant-Pathogenic Fungi. Annu. Rev. Phytopathol. 2014, 52, 427–451.
- Rizo, J.; Südhof, T.C. The Membrane Fusion Enigma: SNAREs, Sec1/Munc18 Proteins, and Their Accomplices—Guilty as Charged? Annu. Rev. Cell Dev. Biol. 2012, 28, 279–308.
- Grissom, J.H.; Segarra, V.A.; Chi, R.J. New Perspectives on Snare Function in the Yeast Minimal Endomembrane System. Genes 2020, 11, 899.
- Rossi, G.; Lepore, D.; Kenner, L.; Czuchra, A.B.; Plooster, M.; Frost, A.; Munson, M.; Brennwald, P. Exocyst Structural Changes Associated with Activation of Tethering Downstream of Rho/Cdc42 GTPases. J. Cell Biol. 2020, 219.
- Zhu, Y.; McFarlane, H.E. Regulation of Cellulose Synthesis via Exocytosis and Endocytosis. Curr. Opin. Plant Biol. 2022, 69, 102273.
- Fasshauer, D.; Jahn, R. Budding Insights on Cell Polarity. Nat. Struct. Mol. Biol. 2007, 14, 360–362.
- Ma, M.; Burd, C.G. Retrograde Trafficking and Quality Control of Yeast Synaptobrevin, Snc1, Are Conferred by Its Transmembrane Domain. Mol. Biol. Cell 2019, 30, 1729–1742.
- Fiedler, M.R.; Cairns, T.C.; Koch, O.; Kubisch, C.; Meyer, V. Conditional Expression of the Small GTPase ArfA Impacts Secretion, Morphology, Growth, and Actin Ring Position in Aspergillus Niger. Front. Microbiol. 2018, 9, 878.
- Burston, H.E.; Maldonado-Báez, L.; Davey, M.; Montpetit, B.; Schluter, C.; Wendland, B.; Conibear, E. Regulators of Yeast Endocytosis Identified by Systematic Quantitative Analysis. J. Cell Biol. 2009, 185, 1097–1110.
- Kashikuma, R.; Nagano, M.; Shimamura, H.; Nukaga, K.; Katsumata, I.; Toshima, J.Y.; Toshima, J. Role of Phosphatidylserine in the Localization of Cell Surface Membrane Proteins in Yeast. Cell Struct. Funct. 2023, 48, 19–30.
- Oberhofer, E. Ubiquitin Modifikation Der Synaptobervine SNC1 Und SNC2 in Saccharomyces Cerevisiae. Bachelor’s Thesis, LMU Munich, Munich, Germany, 2004.
- Burri, L.; Lithgow, T. A Complete Set of SNAREs in Yeast. Traffic 2004, 5, 45–52.
- Lewis, M.J.; Pelham, H.R. A New Yeast Endosomal SNARE Related to Mammalian Syntaxin 8. Traffic 2002, 3, 922–929.
- Shanks, S.G.; Carpp, L.N.; Struthers, M.S.; McCann, R.K.; Bryant, N.J. The Sec1/Munc18 Protein Vps45 Regulates Cellular Levels of Its SNARE Binding Partners Tlg2 and Snc2 in Saccharomyces Cerevisiae. PLoS ONE 2012, 7, e49628.
- Apel, A.R.; Hoban, K.; Chuartzman, S.; Tonikian, R.; Sidhu, S.; Schuldiner, M.; Wendland, B.; Prosser, D. Syp1 Regulates the Clathrin-Mediated and Clathrin-Independent Endocytosis of Multiple Cargo Proteins through a Novel Sorting Motif. Mol. Biol. Cell 2017, 28, 2434–2448.
- Wrasman, K.M. Endocytic Regulation from Cargo to Coat: The Identification of Novel Factors and Endocytic Protein Interactions; Johns Hopkins University: Baltimore, MD, USA, 2016.
- Grote, E.; Vlacich, G.; Pypaert, M.; Novick, P.J. A Snc1 Endocytosis Mutant: Phenotypic Analysis and Suppression by Overproduction of Dihydrosphingosine Phosphate Lyase. Mol. Biol. Cell 2000, 11, 4051–4065.
- Yang, B.; Gonzalez, L.; Prekeris, R.; Steegmaier, M.; Advani, R.J.; Scheller, R.H. SNARE Interactions Are Not Selective: Implications for Membrane Fusion Specificity. J. Biol. Chem. 1999, 274, 5649–5653.
- Furukawa, N.; Mima, J. Multiple and Distinct Strategies of Yeast SNAREs to Confer the Specificity of Membrane Fusion. Sci. Rep. 2014, 4, 4277.
- Yuan, Y.; Zhang, M.; Li, J.; Yang, C.; Abubakar, Y.S.; Chen, X.; Zheng, W.; Wang, Z.; Zheng, H.; Zhou, J. The Small GTPase FgRab1 Plays Indispensable Roles in the Vegetative Growth, Vesicle Fusion, Autophagy and Pathogenicity of Fusarium Graminearum. Int. J. Mol. Sci. 2022, 23, 895.
- Chen, X.; Selvaraj, P.; Lin, L.; Fang, W.; Wu, C.; Yang, P.; Zhang, J.; Abubakar, Y.S.; Yang, F.; Lu, G. Rab7/Retromer-Based Endolysosomal Trafficking Facilitates Effector Secretion and Host Invasion in Rice Blast. bioRxiv 2022.
- Gerst, J.E.; Rodgers, L.; Riggs, M.; Wigler, M. SNC1, a Yeast Homolog of the Synaptic Vesicle-Associated Membrane Protein/Synaptobrevin Gene Family: Genetic Interactions with the RAS and CAP Genes. Proc. Natl. Acad. Sci. USA 1992, 89, 4338–4342.
- Zheng, W.; Lin, Y.; Fang, W.; Zhao, X.; Lou, Y.; Wang, G.; Zheng, H.; Liang, Q.; Abubakar, Y.S.; Olsson, S. The Endosomal Recycling of FgSnc1 by FgSnx41–FgSnx4 Heterodimer Is Essential for Polarized Growth and Pathogenicity in Fusarium Graminearum. New Phytol. 2018, 219, 654–671.
- Valkonen, M. Functional Studies of the Secretory Pathway of Filamentous Fungi: The Effect of Unfolded Protein Response on Protein Production; Edita Prima Oy: Helsinki, Finland, 2003.
- Protopopov, V.; Govindan, B.; Novick, P.; Gerst, J.E. Homologs of the Synaptobrevin/VAMP Family of Synaptic Vesicle Proteins Function on the Late Secretory Pathway in S. Cerevisiae. Cell 1993, 74, 855–861.
- Shen, D.; Yuan, H.; Hutagalung, A.; Verma, A.; Kümmel, D.; Wu, X.; Reinisch, K.; McNew, J.A.; Novick, P. The Synaptobrevin Homologue Snc2p Recruits the Exocyst to Secretory Vesicles by Binding to Sec6p. J. Cell Biol. 2013, 202, 509–526.
- Gerst, J.E. Conserved α-Helical Segments on Yeast Homologs of the Synaptobrevin/VAMP Family of v-SNAREs Mediate Exocytic Function. J. Biol. Chem. 1997, 272, 16591–16598.
- McGuffin, L.J.; Bryson, K.; Jones, D.T. The PSIPRED Protein Structure Prediction Server. Bioinformatics 2000, 16, 404–405.
- Wu, C.; Chen, H.; Yuan, M.; Zhang, M.; Abubakar, Y.S.; Chen, X.; Zhong, H.; Zheng, W.; Zheng, H.; Zhou, J. FgAP1σ Is Critical for Vegetative Growth, Conidiation, Virulence, and DON Biosynthesis in Fusarium Graminearum. J. Fungi 2023, 9, 145.
- Buwa, N.; Martínez-Núñez, L.; Munson, M. Exocytosis in Yeast: Major Players and Mechanisms. In Exocytosis: From Molecules to Cells; IOP Publishing: Bristol, England, 2022.
- Jahn, R.; Scheller, R.H. SNAREs—Engines for Membrane Fusion. Nat. Rev. Mol. Cell Biol. 2006, 7, 631–643.
- Lee, C.; Lepore, D.; Munson, M.; Yoon, T.-Y. Exocyst Stimulates Each Step of Exocytic SNARE Complex Assembly and Vesicle Fusion. bioRxiv 2022.
- Thattai, M. Molecular and Cellular Constraints on Vesicle Traffic Evolution. Curr. Opin. Cell Biol. 2023, 80, 102151.
- Starr, T.L.; Gonçalves, A.P.; Meshgin, N.; Glass, N.L. The Major Cellulases CBH-1 and CBH-2 of Neurospora Crassa Rely on Distinct ER Cargo Adaptors for Efficient ER-exit. Mol. Microbiol. 2018, 107, 229–248.
- Valdez-Taubas, J.; Pelham, H.R. Slow Diffusion of Proteins in the Yeast Plasma Membrane Allows Polarity to Be Maintained by Endocytic Cycling. Curr. Biol. 2003, 13, 1636–1640.
- Bonifacino, J.S.; Rojas, R. Retrograde Transport from Endosomes to the Trans-Golgi Network. Nat. Rev. Mol. Cell Biol. 2006, 7, 568–579.
- Gurunathan, S.; Marash, M.; Weinberger, A.; Gerst, J.E. T-SNARE Phosphorylation Regulates Endocytosis in Yeast. Mol. Biol. Cell 2002, 13, 1594–1607.
- Xu, P.; Hankins, H.M.; MacDonald, C.; Erlinger, S.J.; Frazier, M.N.; Diab, N.S.; Piper, R.C.; Jackson, L.P.; MacGurn, J.A.; Graham, T.R. COPI Mediates Recycling of an Exocytic SNARE by Recognition of a Ubiquitin Sorting Signal. Elife 2017, 6, e28342.
- Zhang, F.; Zhao, M.; Braun, D.R.; Ericksen, S.S.; Piotrowski, J.S.; Nelson, J.; Peng, J.; Ananiev, G.E.; Chanana, S.; Barns, K. A Marine Microbiome Antifungal Targets Urgent-Threat Drug-Resistant Fungi. Science 2020, 370, 974–978.
- Abenza, J.F.; Pantazopoulou, A.; Rodríguez, J.M.; Galindo, A.; Penalva, M.A. Long-distance Movement of Aspergillus Nidulans Early Endosomes on Microtubule Tracks. Traffic 2009, 10, 57–75.
- Hervás-Aguilar, A.; Peñalva, M.A. Endocytic Machinery Protein SlaB Is Dispensable for Polarity Establishment but Necessary for Polarity Maintenance in Hyphal Tip Cells of Aspergillus Nidulans. Eukaryot. Cell 2010, 9, 1504–1518.
- Taheri-Talesh, N.; Horio, T.; Araujo-Bazán, L.; Dou, X.; Espeso, E.A.; Penalva, M.A.; Osmani, S.A.; Oakley, B.R. The Tip Growth Apparatus of Aspergillus Nidulans. Mol. Biol. Cell 2008, 19, 1439–1449.
- Higuchi, Y.; Shoji, J.; Arioka, M.; Kitamoto, K. Endocytosis Is Crucial for Cell Polarity and Apical Membrane Recycling in the Filamentous Fungus Aspergillus Oryzae. Eukaryot. Cell 2009, 8, 37–46.




