Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | MARIA EVANGELINA CAREZZANO | -- | 3292 | 2023-06-06 15:22:29 | | | |
| 2 | Jason Zhu | Meta information modification | 3292 | 2023-06-08 03:55:30 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Carezzano, M.E.; Paletti Rovey, M.F.; Cappellari, L.D.R.; Gallarato, L.A.; Bogino, P.; Oliva, M.D.L.M.; Giordano, W. Biofilm-Forming Ability of Phytopathogenic Bacteria. Encyclopedia. Available online: https://encyclopedia.pub/entry/45246 (accessed on 07 February 2026).
Carezzano ME, Paletti Rovey MF, Cappellari LDR, Gallarato LA, Bogino P, Oliva MDLM, et al. Biofilm-Forming Ability of Phytopathogenic Bacteria. Encyclopedia. Available at: https://encyclopedia.pub/entry/45246. Accessed February 07, 2026.
Carezzano, María Evangelina, María Fernanda Paletti Rovey, Lorena Del Rosario Cappellari, Lucas Antonio Gallarato, Pablo Bogino, María De Las Mercedes Oliva, Walter Giordano. "Biofilm-Forming Ability of Phytopathogenic Bacteria" Encyclopedia, https://encyclopedia.pub/entry/45246 (accessed February 07, 2026).
Carezzano, M.E., Paletti Rovey, M.F., Cappellari, L.D.R., Gallarato, L.A., Bogino, P., Oliva, M.D.L.M., & Giordano, W. (2023, June 06). Biofilm-Forming Ability of Phytopathogenic Bacteria. In Encyclopedia. https://encyclopedia.pub/entry/45246
Carezzano, María Evangelina, et al. "Biofilm-Forming Ability of Phytopathogenic Bacteria." Encyclopedia. Web. 06 June, 2023.
Copy Citation
Phytopathogenic bacteria not only affect crop yield and quality but also the environment. Understanding the mechanisms involved in their survival is essential to develop new strategies to control plant disease. One such mechanism is the formation of biofilms; i.e., microbial communities within a three-dimensional structure that offers adaptive advantages, such as protection against unfavorable environmental conditions. Biofilm-producing phytopathogenic bacteria are difficult to manage. They colonize the intercellular spaces and the vascular system of the host plants and cause a wide range of symptoms such as necrosis, wilting, leaf spots, blight, soft rot, and hyperplasia.
biotic stress
biofilm
phytopathogenic bacteria
1. Biofilm: Composition, Functions, and Stages of Formation
Biofilm has garnered significant scientific interest in the last decade since 99% of the bacterial population can produce it at some point in their life cycle [1]. Biofilms are microbial communities inside structures of their own making, which can adhere to living or inert surfaces. About 10–25% of a biofilm consists of bacterial cells that can belong to members of the same species (in which case the biofilm is simple) or different species (mixed biofilm). The remaining 75–90% is made up of extracellular polymeric substances (EPS), which stabilize and give shape to the matrix [1][2]. This matrix confers enhanced protection to the cells within it against phagocytosis, harmful environmental conditions (pH, lack of nutrients, and mechanical forces), and antibiotics or antimicrobials: in fact, bacteria within a biofilm may be a thousand times more resistant to these agents [1][2][3]. Channels engineered on the inside, moreover, facilitate the circulation and exchange of water, nutrients, and enzymes as well as greater metabolic cooperation between members and the elimination of toxic metabolites [4].
Biofilm formation is a rapid, complex, and dynamic process that depends on changes in the cellular phenotype [5]. Its progressive stages may be summarized as follows:
-
Adhesion: the microorganisms engage in weak interactions (acid-base, hydrophobic, Van der Waals, and electrostatic forces) to reversibly adhere to a surface.
-
Colonization: irreversible bonds come about through hydrophilic/hydrophobic interactions; the bacteria use flagella, pili, and collagen-binding adhesive proteins.
-
Development: EPS are secreted and there is a continuous proliferation and accumulation of cells.
-
Maturation: the three-dimensional structure settles into its stable form featuring circulation and signaling channels.
Certain bacteria within an established biofilm can evolve into persistent cells. These are genetically similar but physiologically different from the parent or primary cells (those that originally colonized the surface). Persistent cells are important in terms of resistance; their metabolism is inert, their replication is slow, and they regulate DNA-repair systems and antitoxin systems [7].
Since biofilm formation is cooperative, it would not be possible without quorum sensing (QS). This bacterium-to-bacterium communication system regulates not only the production of toxins, enzymes, biofilms, and EPS but also virulence factors and infectious processes in pathogenic bacteria [8][9] takes place through the synthesis of low molecular weight signaling molecules known as autoinducers (AI), which act as indicators of population density. When the concentration of these molecules exceeds a certain threshold, they are sensed by receptor molecules that are also synthesized by bacteria, and specific genes are activated (such as those responsible for biofilm production). In short, QS helps bacteria organize themselves into a community through a unified response and, thus, enhances their chances of survival [10].
The regulation of this communication system is very complex and varies from one bacterial species to another. Some of the molecules known to be involved are acyl-homoserine lactones (AHL) in gram-negative bacteria, autoinducing peptides (AIP) in gram-positive bacteria, and autoinducers 2 and 3 (AI-2 and AI-3) in both [5][9][11].
More specifically, QS in most gram-negative bacteria is regulated by a LuxI-LuxR-type system that becomes transcriptionally activated at a certain concentration of extracellularly diffused AHLs. Genes associated with biofilm formation are subsequently expressed. Some gram negatives, such as X. campestris and R. solanacearum, can also synthesize a diffusible signal factor (DSF) as an AI [12].
In gram-positive bacteria, it is a small oligopeptide (a mature AIP) that is produced and then expelled from the cell. Increasing concentrations of this peptide allow it to bind to a histidine kinase enzyme. A phosphorylation cascade ensues and genes related to biofilm formation are expressed [13].
In phytopathogenic bacteria, a global second messenger called cyclic di-GMP (cyclic guanosine monophosphate, abbreviated as cGMP or c-di-GMP) is involved in biofilm formation as well [14]: it regulates EPS biosynthesis and the transition from a mobile planktonic state to one of aggregation. Other processes regulated by this molecule are virulence, the cell cycle, and cell differentiation [8].
2. Social Behavior of the Bacterial Population in the Biofilm Matrix and Its Relationship with Pathogenicity
Social interactions, a common feature within the prokaryotic world [15], make it possible for bacterial populations to respond to environmental variations dynamically and collectively [16]. This adaptive behavior depends on changes at the level of gene expression, which are a result of chemical information in the form of diffusible signal molecules being produced and detected through QS (see Section 4). Moreover, many bacterial species that live in association with plants do so as members of polymicrobial communities, which are self-organized into highly complex biological structures [1][17].
These concepts are the basis of sociomicrobiology, i.e., the study of microbial social behavior. Seen from this perspective, bacteria within a group can act in their self-interest, cooperatively, or with altruism. They may compete against each other, divide labor amongst themselves, or function as donors/recipients in the transfer of genetic material. The biofilm matrix particularly accentuates cooperation and competition. The former occurs when the amount of biofilm formed by an individual species is less than the sum of all the biofilms formed by bacteria from the same environment. The latter is due to the amount of biofilm by one species surpassing the total production by bacteria from different environments [17][18].
In nature, most microorganisms are part of biofilms established as multispecies consortia. This makes sense, given that mixed biofilms tend to have a larger biomass and be more resistant [19]. The dynamics inside these biofilms are probably finely regulated through intra- and inter-species signaling, and members unable to synthesize QS molecules may effectively detect those produced by others [1][20].
Phytopathogenic bacteria within a biofilm can thus act in a coordinated manner to survive, outcompete other microbes, persist in nature, colonize host plants, and eventually infect them. All these processes are far more challenging for bacteria attempting them individually. Biofilms, then, contribute significantly to pathogenicity. In fact, some pathogenic microorganisms can only regulate and express their virulence if environmental conditions and cell density are optimal. Put otherwise, their pathogenicity relies on QS-mediated social activity. For example, P. syringae pv. actinidiae, the causative agent of bacterial canker in kiwi (see also 7.2.), colonizes the plant phyllosphere saprophytically and only penetrates the plant through wounds and natural openings when bacterial cell density is suitable [21]. Therefore, the study of these ecological relationships is a topic of increasing interest for the understanding of plant pathologies [22][23].
3. Biofilm in Plants
Biofilms were first observed on leaf surfaces in the early 1960s and on roots in the 1970s. Since then, evidence has accumulated of plant-associated bacterial aggregates, microcolonies, and biofilms. Plants harbor a diverse bacterial community on leaves, roots, shoots, and/or within their tissues. Bacteria can even be found in the depressions at the junctions of epidermal cells.
Plant-bacteria interactions can either harm or benefit plant health and yield. In other words, some biofilm-forming bacteria boost plant growth and protect against disease, while others are phytopathogenic [8][24][25]. For example, plant-growth-promoting rhizobacteria (PGPR) ensure effective root colonization by forming a biofilm, and in so doing enhance the plants’ ability to synthesize useful hormones and acquire nutrients, their resistance to stress, and, therefore, their yield [26][27]. In contrast, the biofilms formed in vascular systems by pathogenic bacteria, such as R. solanacearum, Xyllela fastidiosa, and Pantoea stewartii, cause wilting [24][28]. Certain human biofilm-producing pathogens, such as P. aeruginosa, can also infect the roots of plants, such as Arabidopsis, by producing the same virulence factors [28].
The ability of a pathogenic bacterium to successfully produce a biofilm on a plant host is strongly influenced by plant-microorganism interactions, the environment, and the host’s own immune response, physiological status, nutritional status, and signaling system, as described in Section 5 [8].
4. Biofilm Formed by Phytopathogenic Bacteria
The impact of phytopathogenic biofilms on agriculture cannot be understated. Formed on leaves (mesophyll, parenchyma), in the rhizosphere, and/or in vascular bundles [29][30], they reduce crop yield and quality and affect the safety of agricultural products intended for human consumption and animal feeding. In addition, the EPSs secreted by phytopathogenic bacteria interfere with the proper functioning of plant tissues and organs [29]. All of this seriously undermines food security at a time when booming populations around the world require high productivity rates, as indicated by the UN [26].
The current strategies to control biofilms include pesticides and antibiotics, but they are not efficient enough and they pose their own risks. Infections usually reappear and the overuse of chemical products causes water and soil contamination. If new management strategies are to emerge, they will demand thorough knowledge of how phytopathogenic biofilm is produced and regulated [11].
Several studies have focused on the importance of biofilm formation in plant pathogenesis. Many phytopathogenic bacteria produce biofilm on the leaf surface, such as P. syringae pv. theae, which is able to survive drought by living within biofilms on tea leaves [31]. Other Pseudomonas spp., such as P. aeruginosa on Arabidopsis taliana and P. syringae pv. syringae B728a, form biofilms on trichomes. These structures retain water and contain nutrients, two key components for the creation and endurance of the 3-D matrix [32]. Biofilm may be formed in xylem vessels and roots as well (see following subsections). Other examples of biofilm-producing phytopathogenic bacteria are A. tumefaciens, Xylella fastidiosa, Erwinia amylovora, P. stewartii, R. solanacearum, and X. campestris [8][33].
The disease cycle, which comprises differentiated stages and is represented in Figure 1, starts with a source of inoculum and ends with the appearance of symptoms such as tumors [34], decay and chlorosis [35], blight [36], wilting [37], rot [38], and cankers [39].
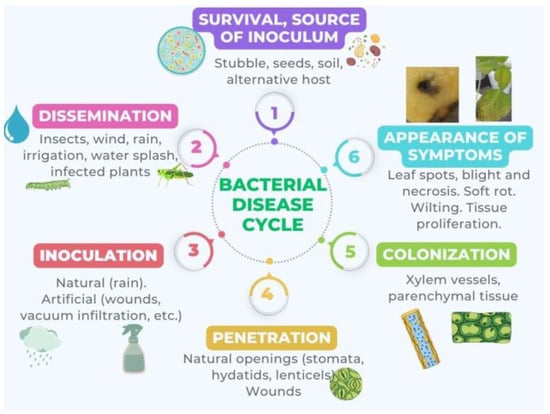
Figure 1. Cyclic model of disease by phytopathogenic bacteria consisting of several distinct stages: 1. Survival or source of inoculum: The bacteria survive outside the host by developing in another environment or by remaining dormant. 2. Dissemination: The bacteria spread. 3. Inoculation: The bacteria come into contact with the plant. 4. Penetration: The bacteria enter the plant. 5. Colonization: The bacteria disseminate within the plant. 6. Appearance of symptoms: This is the result of bacteria producing phytotoxins, EPS, exoenzymes, phytohormones, etc.
QS is responsible for regulating pathogenicity and colonization [9][40], and some phytopathogenic bacteria may have more than one QS system (featuring AHLs or diffusible signal factors, DSF) as well as a virulence factor modulation system [41]. For bacteria such as E. amylovora, P. syringae, Xanthomonas spp., and Ralstonia spp., T3SS are also involved in pathogenesis since they enable the direct introduction of pathogenic proteins into host cells. These systems are encoded by hypersensitivity response and pathogenicity (hrp) genes, which are classified into two groups with different organizations and modes of regulation. The hrp in group 1, typical of Pseudomonas, Erwinia, and Pantoea spp., are activated by complex regulatory pathways that end in proteins HrpS and HrpL. Xanthomonas and Ralstonia spp., on the other hand, have group II hrp [42].
4.1. Phytopathogenic Bacteria that Colonize Xylem Vessels
The causative agent of fire blight, E. amylovora, colonizes rosaceous plants by regulating their immune responses and physiology through a T3SS. In addition, it produces two EPS, amylovoran and levan, to form a biofilm within the vascular tissue. The synthesis of these polymers and that of cellulose, another component of the biofilm matrix, are positively regulated by an increase in intracellular c-di-GMP [8][43].
Gram-negative bacteria belonging to the genus Dickeya (formerly Erwinia) cause soft rot by synthesizing pectinase, an enzyme that degrades pectin in the cell wall and the middle lamella. The process is known to be regulated by QS and virulence factor modulation [44]. D. dadantii, responsible for stem and root rot in sweet potatoes, grows in biofilms and regulates the colonization of intercellular spaces and xylem vessels through flagella-mediated motility, the synthesis of LPS and extracellular polysaccharides, biofilm formation, and a T3SS [45][46]. Another example within the same genus is D. zeae, which affects economically important crops such as maize, bananas, rice, and potatoes [47].
Pectobacterium carovotum (also formerly classified as an Erwinia, but now within the genus Pectobacterium) forms a biofilm in the xylems of susceptible host plants, such as potatoes [8]. Different strains vary in their virulence depending on the concentrations of AHLs, which once again shows how crucial QS is to successful infection. Much like Dickeya spp., P. carovotum hydrolyzes pectin between plant cells and brings about soft rot. In addition, it can “hijack” the host’s genes to foster the development of disease [48][49].
Pierce’s disease in vines and variegated chlorosis in citrus plants occur when extensive biofilms of X. fastidiosa are created in the vascular system. In those areas where biofilm manages to block the nutrient flow, visible symptoms appear. Grasshoppers act as vectors: the bacterium colonizes their large intestine thanks to a QS system whose signal molecules are DSF and rpf gene products. When X. fastidiosa senses that its population density within the plant is high, the synthesis of c-di-GMP is inhibited and the formation of EPS and biofilm in the xylem are promoted. Conversely, low density is associated with an increase in intracellular c-di-GMP and the inhibition of adhesion and biofilm formation. This makes it possible for the pathogen to circulate freely through the plant and to be transferred into the insect vectors that feed on the plant’s sap [8][28][50][51].
X. campestris pv. campestris, the causal agent of black rot in crucifers, colonizes the xylem after gaining access to the plant through wounds or hydathodes. It synthesizes xanthan gum to form biofilm and degrading exoenzymes that promote virulence. Its aggregation is regulated by c-di-GMP and a two-component RpfC/RpfG system, in which RpfC is the histidine kinase sensor and RpfG is the response-regulating protein. When intracellular c-di-GMP is high, a protein similar to the cyclic AMP Clp receptor changes its conformation and cannot bind to target sites such as the promoter region in manA, which codes for endomannanase, a biofilm-dispersing enzyme. Genes in the xag cluster, on the other hand, code for a glycosyl transferase that is important for biofilm formation [8][50].
Sweet corn and maize may suffer from Stewart’s wilt, a disease transmitted by the corn flea beetle and caused by P. stewartii subsp. stewartii. Through the intervention of an hrp-encoded Hrp T3SS and the effector WtsE, the bacterium infects the apoplast and the xylem. There, its population density grows and dense biofilms are formed, encapsulated in a slime exopolysaccharide called stewartan. The water flow is blocked and symptoms appear, ranging from chlorotic lesions on leaves that eventually become necrotic and delay growth to rapid wilting and death in more susceptible plants. Stewartan also facilitates the pathogen’s movement through the vessels or intercellular spaces, which increases virulence [42][50][52][53].
Fatal wilt in more than 200 plant species is caused by several R. solanacearum strains that live in the soil, preferably in the deeper layers. Water and infected weeds can function as its reservoirs. It infects the roots and travels to the xylem where it multiplies, obstructs the vessels with large amounts of EPS, and, in the end, causes the plant to wither and die. Other factors implicated in its virulence are the synthesis of wall-degrading enzymes [50][54][55], chemotaxis, and motility. Chemotaxis (as described in Section 3) takes place when receptors in the bacterial cell membrane detect specific chemical substances to which they are attracted. Motility can take different forms depending on the appendages driving it. A rotary movement called swimming is produced through the use of polar flagella, while coordinated multicellular movement can be achieved by extending, attaching, and retracting type IV pili (see Section 4) [56][57].
A gram-positive pathogen that targets xylem vessels is Clavibacter michiganensis. Clavibacter michiganensis subspecies michiganensis infects tomatoes, and Clavibacter michiganensis subspecies sepedonicus gives rise to ring rot in potatoes [50][58][59]. C. michiganensis subsp. michiganensis is the causal agent of bacterial wilt and canker in tomatoes. Unilateral wilting in the host plant during the early stages of infection may mean that the pathogen has invaded the protoxylem, not the adjacent vessels, in which case the plant is still safe as a nutritional source. Dehydration and death ensue when C. michiganensis multiplies and produces EPS and glycoproteins to create large biofilms that decrease the water flow [60][61][62].
4.2. Phytopathogenic Bacteria that Colonize Root Tissues
Dicotyledonous plants suffer serious damage when tumors called “galls” appear in the junction between the roots and the stem. The formation of these tumors is induced by genes inside a Ti plasmid that belongs to a parasitic bacterium, A. tumefaciens, which produces biofilm mainly on the roots. For this purpose, it synthesizes cellulose and a unipolar polysaccharide adhesin (UPP) when intracellular c-di-GMP is high [1][8][63]. Some of the regulatory pathways for biofilm production in A. tumefaciens include an oxygen limitation response pathway, a two-component PhoR-PhoP system (involved in adhesion and biomass increase), and a regulator of ExoR secretion (which has to do with motility) [64].
A soil-borne pathogen that has been reclassified as A. rhizogenes (after being considered a rhizobium) forms large biofilms and causes hairy root disease in hydroponics. It has a signaling system made up of AHLs, and it introduces its DNA into the plant genome to live at the expense of its host [65][66][67].
4.3. Phytopathogenic Bacteria that Colonize Parenchymal Tissues
Phytopathogenic bacteria that form biofilm on leaf surfaces enter the leaf through natural openings called stomata. Many of them synthesize an ice nucleation protein that allows them to survive inside the leaves at low temperatures [68]. Pseudomonas spp. are prominent examples. Their ability to form a biofilm has been shown to depend on the incubation time and the availability of nutrients since different species adopt different strategies for colonization [69]. P. syringae, a widespread pathogen among crops, poses a significant threat to food security worldwide. More than 50 pathovars have been identified; each one with a high degree of host specificity and, therefore, the ability to infect only a limited number of plant species or even a few cultivars of a single species. This specificity is the basis for classifying P. syringae strains into different pathovars (pv.) [70][71]. For instance, the Pseudomonas strain that infects tomato plants has been named P. syringae pv. tomato, and the one responsible for canker in kiwifruit is P. syringae pv. actinidiae. Their production of biofilm on the leaf surface and their virulence are favored by the production of alginate, a polysaccharide [72][73]. This production and that of acetylated cellulose, another important component of their biofilm matrix, is regulated by algU genes, which are additionally involved in osmotolerance and motility. The synthesis of acetylated cellulose is also regulated by a wssABCDEFGHI operon. As with other bacteria reviewed here, c-di-GMP plays its part in biofilm formation [74][75].
P. syringae strains, moreover, use a T3SS to inject plant cells with virulence factors, such as protein effectors and a phytotoxin (coronatin) that mimics the plant hormone methyl jasmonate. This grants bacteria the ability to decrease the host’s immune response and increase its susceptibility to disease [8][76][77]. The T3SS is encoded by a group of hrp and hypersensitive and conserved response (hrc) genes, which are strictly controlled by the enhancer-coding proteins HrpR and HrpS. These proteins cooperatively activate HrpL expression, controlled by sigma-54. The emergence of P. syringae strains that have developed resistance to traditionally used antimicrobials makes their control all the more difficult [78][79]. P. savastanoi pv. glycinea, another Pseudomonas, also uses an hrp-encoded T3SS, coronatin, and biofilm to cause blight in soybean [77][80]. An example outside the Pseudomonas genus (although it was formerly classified within it) is Acidovorax citrulli, the causative agent of fruit spots in cucurbits. Its pathogenicity is not only related to biofilm formation but also to a T6SS, an R3SS, and the use of QS [81][82].
References
- Bogino, P.; Abod, A.; Nievas, F.; Giordano, W. Water-limiting conditions alter the structure and biofilm-forming ability of bacterial multispecies communities in the alfalfa rhizosphere. PLoS ONE 2013, 8, e79614.
- Liaqat, I.; Liaqat, M.; Tahir, H.M.; Haq, I.; Ali, N.M.; Arshad, M.; Arshad, N. Motility effects biofilm formation in Pseudomonas aeruginosa and Enterobacter cloacae. Pak. J. Pharm. Sci. 2019, 32, 927–932.
- Beoletto, V.G.; De Las Mercedes Oliva, M.; Marioli, J.M.; Carezzano, M.E.; Demo, M.S. Antimicrobial natural products against bacterial biofilms. In Antibiotic Resistance: Mechanisms and New Antimicrobial Approaches; Kon, K., Rai, M., Eds.; Elsevier: London, UK, 2016; pp. 290–307.
- Srinivasan, R.; Santhakumari, S.; Poonguzhali, P.; Geetha, M.; Dyavaiah, M.; Xiangmin, L. Bacterial biofilm inhibition: A focused review on recent therapeutic strategies for combating the biofilm mediated infections. Front. Microbiol. 2021, 12, 676458.
- Masák, J.; Čejková, A.; Schreiberová, O.; Rezanka, T. Pseudomonas biofilms: Possibilities of their control. FEMS Microbiol. Ecol. 2014, 89, 1–14.
- Yin, W.; Wang, Y.; Liu, L.; He, J. Biofilms: The microbial “protective clothing” in extreme environments. Int. J. Mol. Sci. 2019, 20, 3423.
- Kumar, A.; Alam, A.; Rani, M.; Ehtesham, N.Z.; Hasnain, S.E. Biofilms: Survival and defense strategy for pathogens. Int. J. Med. Microbiol. 2017, 307, 481–489.
- Castiblanco, L.F.; Sundin, G.W. New insights on molecular regulation of biofilm formation in plant-associated bacteria. J. Integr. Plant Biol. 2016, 58, 362–372.
- Baltenneck, J.; Reverchon, S.; Hommais, F. Regulación de detección de quórum en bacterias fitopatógenas. Microorganismos 2021, 9, 239.
- Brindhadevi, K.; LewisOscar, F.; Mylonakis, E.; Shanmugam, S.; Verma, T.N.; Pugazhendhi, A. Biofilm and quorum sensing mediated pathogenicity in Pseudomonas aeruginosa. Process Biochem. 2020, 96, 49–57.
- Preda, V.G.; Săndulescu, O. Communication is the key: Biofilms, quorum sensing, formation and prevention. Discoveries 2019, 7, e100.
- Li, J.; Zhao, X. Effects of quorum sensing on the biofilm formation and viable but non-culturable state. Food Res. Int. 2020, 137, 109742.
- Zhou, L.; Zhang, Y.; Ge, Y.; Zhu, X.; Pan, J. Regulatory mechanisms and promising applications of quorum sensing-inhibiting agents in control of bacterial biofilm formation. Front. Microbiol. 2020, 11, 589640.
- Satpathy, S.; Kumar Sen, S.; Pattanaik, S.; Raut, S. Review on bacterial biofilm: An universal cause of contamination. Biocatal. Agric. Biotechnol. 2016, 7, 56–66.
- Whiteley, M.; Diggle, S.; Greenberg, E. Progress in and promise of bacterial quorum sensing research. Nature 2017, 551, 313–320.
- Atkinson, S.; Williams, P. Quorum sensing and social networking in the microbial world. J. R. Soc. Interface 2009, 6, 959–978.
- Parsek, M.R.; Greenberg, E.P. Sociomicrobiology: The connections between quorum sensing and biofilms. Trends Microbiol. 2005, 13, 27–33.
- Zhou, J.; Cai, Z. Microbial Social Interactions in Biofilm. In Implication of Quorum Sensing System in Biofilm Formation and Virulence; Bramhachari, P.V., Ed.; Springer: Berlin/Heidelberg, Germany, 2018; pp. 29–46.
- Velmourougane, K.; Prasanna, R.; Saxena, A.K. Agriculturally important microbial biofilms: Present status and future prospects. J. Basic Microbiol. 2017, 57, 548–573.
- Diggle, S.P.; Gardner, A.; West, S.A.; Griffin, A.S. Evolutionary theory of bacterial quorum sensing: When is a signal not a signal? Philos. Trans, R. Soc. Lond B. Biol. Sci. 2007, 362, 1241–1249.
- Cellini, A.; Donati, I.; Fiorentini, L.; Vandelle, E.; Polverari, A.; Venturi, V.; Buriani, G.; Vanneste, J.L.; Spinelli, F. N-Acyl Homoserine lactones and lux solos regulate social behaviour and virulence of Pseudomonas syringae pv. actinidiae. Microb. Ecol. 2020, 79, 383–396.
- Nadell, C.D.; Xavier, J.B.; Foster, K.R. The sociobiology of biofilms. FEMS Microbiology Reviews 2009, 33, 206–224.
- Li, Y.; Tian, X. Quorum sensing and bacterial social interactions in biofilms. Sensors 2012, 12, 2519–2538.
- Morris, C.E.; Monier, J.M. The ecological significance of biofilm formation by plant-associated bacteria. Annu. Rev. Phytopathol. 2003, 41, 429–453.
- Patwardhan, S.B.; Pandit, C.; Pandit, S.; Verma, D.; Lahiri, D.; Nag, M.; Ray, R.R.; Jha, P.; Prasad, R. Illuminating the signalomics of microbial biofilm on plant surfaces, Biocatal. Agric. Biotechnol. 2023, 47, 102537.
- Rafique, M.; Hayat, K.; Mukhtar, T.; Khan, A.A.; Afridi, M.S.; Hussain, T.; Sultan, T.; Munis, M.F.H.; Imran, M.; Chaudhary, H.J. Bacterial Biofilm Formation and Its Role Against Agricultural Pathogens. In The Battle against Microbial Pathogens: Basic Science, Technological Advances and Educational Programs; Mendez Vilas, A., Ed.; Formatex Research Center: Badajoz, Spain, 2015; pp. 373–382.
- Ajijah, N.; Fiodor, A.; Pandey, A.K.; Rana, A.; Pranaw, K. Plant Growth-Promoting Bacteria (PGPB) with biofilm-forming ability: A multifaceted agent for sustainable agriculture. Diversity 2023, 15, 112.
- Rudrappa, T.; Bais, H.P. Curcumin, a known phenolic from Curcuma longa, attenuates the virulence of Pseudomonas aeruginosa PAO1 in whole plant and animal pathogenicity models. J. Agric. Food Chem. 2008, 56, 1955–1962.
- Mina, I.R.; Jara, N.P.; Criollo, J.E.; Castillo, J.A. The critical role of biofilms in bacterial vascular plant pathogenesis. Plant Pathol. 2019, 68, 1439–1447.
- Zhu, X.; Chen, W.J.; Bhatt, K.; Zhou, Z.; Huang, Y.; Zhang, L.H.; Chen, S.; Wang, J. Innovative microbial disease biocontrol strategies mediated by quorum quenching and their multifaceted applications: A review. Front. Plant Sci. 2023, 13, 1063393.
- Tomihama, T.; Nishi, Y.; Arai, K. Biofilm formation and resistance to bactericides of Pseudomonas syringae pv. theae. J. Gen. Plant Pathol. 2006, 73, 193–196.
- Lomovatskaya, L.A.; Romanenko, A.S. Secretion systems of bacterial phytopathogens and mutualists (Review). Appl. Biochem. Microbiol. 2020, 56, 115–129.
- Lauber, F.; Deme, J.C.; Lea, S.M.; Berks, B.C. Type 9 secretion system structures reveal a new protein transport mechanism. Nature 2018, 564, 77–82.
- Sidorova, D.E.; Skripka, M.I.; Khmel, I.A.; Koksharova, O.A.; Plyuta, V.A. Effects of volatile organic compounds on biofilms and swimming motility of Agrobacterium tumefaciens. Microorganisms 2022, 10, 1512.
- Zecharia, N.; Krasnov, H.; Vanunu, M.; Siri, A.C.; Haberman, A.; Dror, O.; Vakal, L.; Almeida, R.P.P.; Blank, L.; Shtienberg, D.; et al. Xylella fastidiosa Outbreak in Israel: Population genetics, host range, and temporal and spatial distribution analysis. Phytopathology 2022, 11, 2296–2309.
- Dagher, F.; Nickzad, A.; Zheng, J.; Hoffmann, M.; Déziel, E. Characterization of the biocontrol activity of three bacterial isolates against the phytopathogen Erwinia amylovora. Microbiology 2021, 10, e1202.
- Sowndarya, J.; Rubini, D.; Sinsinwar, S.; Senthilkumar, M.; Nithyanand, P.; Vadivel, V. Gallic acid an agricultural byproduct modulates the biofilm matrix exopolysaccharides of the phytopathogen Ralstonia solanacearum. Curr. Microbiol. 2020, 77, 3339–3354.
- Cai, Z.; Yuan, Z.-H.; Zhang, H.; Pan, Y.; Wu, Y.; Tian, X.-Q.; Wang, F.-F.; Wang, L.; Qian, W. Fatty acid DSF binds and allosterically activates histidine kinase RpfC of phytopathogenic bacterium Xanthomonas campestris pv. campestris to regulate quorum-sensing and virulence. PLoS Pathog. 2017, 13, e1006304.
- Ndemueda, A.; Pereira, I.; Faustino, M.A.F.; Cunha, Â. Photodynamic inactivation of the phytopathogenic bacterium Xanthomonas citri subsp. citri. Lett. Appl. Microbiol. 2020, 4, 420–427.
- Majdura, J.; Jankiewicz, U.; Gałązka, A.; Orzechowski, S. The role of quorum sensing molecules in bacterial–plant interactions. Metabolites 2023, 13, 114.
- Sibanda, S.; Moleleki, L.N.; Shyntum, D.Y.; Coutinho, T.A. Quorum Sensing in Gram-Negative Plant Pathogenic Bacteria. In Advances in Plant Pathology; Kimatu, J.N., Ed.; IntechOpen: London, UK, 2018; Volume 10.
- Merighi, M.; Majerczak, D.R.; Coplin, D.L. A novel transcriptional autoregulatory loop enhances expression of the Pantoea stewartii subsp. stewartii Hrp type III secretion system. FEMS Microbiol. Lett. 2005, 243, 479–487.
- Yuan, X.; Hulin, M.T.; Sundin, G.W. Effectors, chaperones, and harpins of the Type III secretion system in the fire blight pathogen Erwinia amylovora: A review. J. Plant Pathol. 2021, 103, 25–39.
- Liu, F.; Hu, M.; Zhang, Z.; Xue, Y.; Chen, S.; Hu, A.; Zhang, L.-h.; Zhou, J. Dickeya manipulates multiple quorum sensing systems to control virulence and collective behaviors. Front Plant Sci. 2022, 13, 68.
- Hossain, A.; Abdallah, Y.; Ali, M.A.; Masum, M.M.I.; Li, B.; Sun, G.; Meng, Y.; Wang, Y.; An, Q. Lemon-fruit-based green synthesis of zinc oxide nanoparticles and titanium dioxide nanoparticles against soft rot bacterial pathogen Dickeya dadantii. Biomolecules 2019, 9, 863.
- Hossain, A.; Hong, X.; Ibrahim, E.; Li, B.; Sun, G.; Meng, Y.; Wang, Y.; An, Q. Green synthesis of silver nanoparticles with culture supernatant of a bacterium Pseudomonas rhodesiae and their antibacterial activity against soft rot pathogen Dickeya dadantii. Molecules 2019, 24, 2303.
- Jiang, S.; Zhang, J.; Yang, Q.; Sun, D.; Pu, X.; Shen, H.; Li, Q.; Wang, Z.; Lin, B. Antimicrobial activity of natural plant compound carvacrol against soft rot disease agent Dickeya zeae. Curr. Microbiol. 2021, 78, 3453–3463.
- Hajian-Maleki, H.; Sareh, B.-R.; Mohammad, M. Efficiency of essential oils against Pectobacterium carotovorum subsp. carotovorum causing potato soft rot and their possible application as coatings in storage. Postharvest Biol. Technol. 2019, 156, 110928.
- Li, B.; Huang, J.; Yi, Y.; Liu, S.; Liu, R.; Xiao, Z.; Li, C. Effects of rhapontigenin as a novel quorum-sensing inhibitor on exoenzymes and biofilm formation of Pectobacterium carotovorum subsp. carotovorum and its application in vegetables. Molecules 2022, 27, 8878.
- Danhorn, T.; Fuqua, C. Biofilm formation by plant-associated bacteria. Annu. Rev. Microbiol. 2007, 61, 401–422.
- Feitosa-Junior, O.R.; Souza, A.P.S.; Zaini, P.A.; Baccari, C.; Ionescu, M.; Pierry, P.M.; Uceda-Campos, G.; Labroussaa, F.; Almeida, R.P.P.; Lindow, S.E.; et al. The XadA trimeric autotransporter adhesins in Xylella fastidiosa differentially contribute to cell aggregation, biofilm formation, insect transmission and virulence to plants. Mol. Plant-Microbe Interact. 2022, 35, 857–866.
- Roper, M.C. Pantoea stewartii subsp. stewartii: Lessons learned from a xylem-dwelling pathogen of sweet corn. Mol. Plant Pathol. 2011, 12, 628–637.
- Bartholomew, H.P.; Reynoso, G.; Thomas Brandi, J.; Mullins Chase, M.; Smith, C.; Gentzel, I.N.; Giese, L.A.; Mackey, D.; Stevens, A.M. The Transcription factor Lrp of Pantoea stewartii subsp. stewartii controls capsule production, motility, and virulence important for in planta growth. Front. Microbiol. 2022, 12, 4229.
- Malafaia, C.B.; Jardelino, A.C.S.; Silva, A.G.; de Souza, E.B.; Macedo, A.J.; Correia, M.T.D.S.; Silva, M.V. Effects of caatinga plant extracts in planktonic growth and biofilm formation in Ralstonia solanacearum. Microb. Ecol. 2018, 75, 555–561.
- Pau, S.; de Roger, P.-J.; Benoit, D.; Anurag, K.; Núria, S.C.; Marc, V. The Bacterial wilt reservoir host solanum dulcamara shows resistance to Ralstonia solanacearum Infection. Front. Plant Sci. 2021, 12, 755708.
- Corral, J.; Sebastià, P.; Coll, N.S.; Barbé, J.; Aranda, J.; Valls, M. Twitching and swimming motility play a role in Ralstonia solanacearum pathogenicity. Msphere. 2020, 5, e00740-19.
- Yoshihara, A.; Shimatani, M.; Sakata, M.; Takemura, C.; Senuma, W.; Hikichi, Y.; Kai, K. Quorum sensing inhibition attenuates the virulence of the plant pathogen Ralstonia solanacearum species complex. ACS Chem. Biol. 2020, 15, 3050–3059.
- Eichenlaub, R.; Gartemann, K.H. The Clavibacter michiganensis subspecies: Molecular investigation of gram-positive bacterial plant pathogens. Annu. Rev. Phytopathol. 2011, 49, 445–464.
- Peritore-Galve, F.C.; Miller, C.; Smart, C.D. Characterizing colonization patterns of Clavibacter michiganensis during infection of tolerant wild Solanum species. Phytopathology 2020, 110, 574–581.
- Chalupowicz, L.; Zellermann, E.M.; Fluegel, M.; Dror, O.; Eichenlaub, R.; Gartemann, K.H.; Savidor, A.; Sessa, G.; Iraki, N.; Barash, I.; et al. Colonization and movement of GFP-labeled Clavibacter michiganensis subsp. michiganensis during tomato infection. Phytopathology 2012, 102, 23–31.
- Flügel, M.; Becker, A.; Gartemann, K.H.; Eichenlaub, R. Analysis of the interaction of Clavibacter michiganensis subsp. michiganensis with its host plant tomato by genome-wide expression profiling. J. Biotechnol. 2022, 160, 42–54.
- Rivera-Sosa, L.M.; Ramírez-Valverde, G.; Martínez-Yáñez, B.; Judith-Hernández, A.; Aranda-Ocampo, S. Response of tomato (Solanum lycopersicum) varieties to Clavibacter michiganensis subsp. michiganensis infection. Rev. Mex. Fitopatol. 2022, 40, 18–39.
- Padmavathi, A.R.; Bakkiyaraj, D.; Pandian, S.K. Biochemical and molecular mechanisms in biofilm formation of plant-associated bacteria. In Biofilms in Plant and Soil Health; Ahmad, I., Husain, F.M., Eds.; Wiley: Hoboken, NJ, USA, 2017; pp. 195–214.
- Thompson, M.A.; Onyeziri, M.C.; Fuqua, C. Function and regulation of Agrobacterium tumefaciens cell surface structures that promote attachment. In Agrobacterium Biology. Current Topics in Microbiology and Immunology; Gelvin, S., Ed.; Springer: Cham, Switzerland, 2018; Volume 418, pp. 143–184.
- Kumar Junta, M.; Gupta, A.K.; Mahajan, R. Biological control of hairy root (Rhizobium rhizogenes) in apple nurseries through Rhizobium radiobacter antagonists (strain K-84 and native strain UHFBA-218). Biol. Control. 2021, 164, 104762.
- Bourigault, Y.; Rodrigues, S.; Crépin, A.; Chane, A.; Taupin, L.; Bouteiller, M.; Dupont, C.; Merieau, A.; Konto-Ghiorghi, Y.; Boukerb, A.M.; et al. Biocontrol of biofilm formation: Jamming of sessile-associated rhizobial communication by Rhodococcal Quorum-Quenching. Int. J. Mol. Sci. 2021, 22, 8241.
- Rajkumari, J.; Katiyar, P.; Dheeman, S.; Pandey, P.; Maheshwari, D.K. The changing paradigm of rhizobial taxonomy and its systematic growth upto postgenomic technologies. World J. Microbiol. Biotechnol. 2022, 38, 206.
- Tomihama, T.; Nonaka, T.; Nishi, Y.; Arai, K. Environmental control in tea fields to reduce infection by Pseudomonas syringae pv. theae. Phytopathology 2009, 99, 209–216.
- Ueda, A.; Saneoka, H. Caracterización de la capacidad de formar biopelículas por especies de Pseudomonas asociadas a plantas. Curr. Microbiol. 2015, 70, 506–513.
- Xin, X.F.; He, S.Y. Pseudomonas syringae pv. tomato DC3000: A model pathogen for probing disease susceptibility and hormone signaling in plants. Annu. Rev. Phytopathol. 2013, 51, 473–498.
- Xin, X.F.; Kvitko, B.; He, S.Y. Pseudomonas syringae: What it takes to be a pathogen. Nat. Rev. Microbiol. 2018, 16, 316–328.
- Patyka, V.; Buletsa, N.M.; Pasichnyk, L.A.; Zhitkevich, N.; Kalinichenko, A.; Gnatiuk, T.T.; Butsenko, L.N. Specifics of pesticides effects on the phytopathogenic bacteria. Ecol. Chem. Eng. S. 2016, 23, 311–331.
- Shao, X.; Xie, Y.; Zhang, Y.; Deng, X. Biofilm formation assay in Pseudomonas syringae. Bio. Protoc. 2019, 9, e3237.
- O’Malley, M.R.; Anderson, J.C. Regulation of the Pseudomonas syringae Type III secretion system by host environment signals. Microorganisms 2021, 9, 1227.
- Fishman, M. Signaling Dynamics in Pseudomonas syringae pv. Tomato DC3000. Ph.D. Thesis, Cornell University, Ithaca, NY, USA, 2018.
- Engl, C.; Waite, C.J.; McKenna, J.F.; Bennett, M.H.; Hamann, T.; Buck, M. Chp8, a Diguanylate cyclase from Pseudomonas syringae pv. tomato DC3000, suppresses the pathogen-associated molecular pattern flagellin, increases extracellular polysaccharides, and promotes plant immune evasion. mBio 2014, 5, e01168-14.
- Carezzano, E.; Sotelo, J.; Primo, E.; Reinoso, E.; Paletti Rovey, M.F.; Demo, M.; Giordano, W.; Oliva, M.D.L.M. Inhibitory effect of Thymus vulgaris and Origanum vulgare EO on virulence factors of phytopathogenic Pseudomonas syringae strains. Plant Biol. 2017, 19, 599–607.
- Brunetti, A.; Pucci, N.; Modesti, V.; Lumia, V.; Latini, A.; Loreti, S.; Pilotti, M. In vitro and in planta screening of compounds for the control of Pseudomonas syringae pv. actinidiae in Actinidia chinensis var. chinensis. Eur. J. Plant Pathol. 2020, 158, 829–848.
- Han, Q.; Feng, L.; Zhang, Y.; Zhang, R.; Wang, G.; Zhang, Y. Effect of Juglone against Pseudomonas syringae pv. actinidiae planktonic growth and biofilm formation. Molecules 2021, 26, 7580.
- Nguyen, V.T.; Sakata, N.; Usuki, G.; Ishiga, T.; Hashimoto, Y.; Ishiga, Y. Multiple virulence factors regulated by AlgU contribute to the pathogenicity of Pseudomonas savastanoi pv. glycinea in soybean. PeerJ 2021, 9, e12405.
- Fei, N.; Ji, W.; Yang, L.; Yu, C.; Qiao, P.; Yan, J.; Guan, W.; Yang, Y.; Zhao, T. Hcp of the Type VI Secretion System (T6SS) in Acidovorax citrulli group II strain Aac5 has a dual role as a core structural protein and an effector protein in colonization, growth ability, competition, biofilm formation, and ferric iron absorption. Int. J. Mol. Sci. 2022, 23, 9632.
- Ji, W.; Zhao, M.; Fei, N.; Yang, L.; Qiao, P.; Walcott, R.; Yang, Y.; Zhao, T. Essential Acidovorax citrulli virulence gene hrpE activates host immune response against pathogen. Int. J. Mol. Sci. 2022, 23, 9144.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
851
Revisions:
2 times
(View History)
Update Date:
08 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




