Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Yuzhu Zhang | -- | 3643 | 2023-06-05 19:53:05 | | | |
| 2 | Rita Xu | Meta information modification | 3643 | 2023-06-06 03:45:27 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Zhang, Y.; Che, H.; Li, C.; Jin, T. Food Allergens of Plant Origin. Encyclopedia. Available online: https://encyclopedia.pub/entry/45206 (accessed on 05 March 2026).
Zhang Y, Che H, Li C, Jin T. Food Allergens of Plant Origin. Encyclopedia. Available at: https://encyclopedia.pub/entry/45206. Accessed March 05, 2026.
Zhang, Yuzhu, Huilian Che, Caiming Li, Tengchuan Jin. "Food Allergens of Plant Origin" Encyclopedia, https://encyclopedia.pub/entry/45206 (accessed March 05, 2026).
Zhang, Y., Che, H., Li, C., & Jin, T. (2023, June 05). Food Allergens of Plant Origin. In Encyclopedia. https://encyclopedia.pub/entry/45206
Zhang, Yuzhu, et al. "Food Allergens of Plant Origin." Encyclopedia. Web. 05 June, 2023.
Copy Citation
The structures and structural components of the food allergens in the allergen families may provide further directions for discovering new food allergens. Answers as to what makes some food proteins allergens are still elusive. Factors to be considered in mitigating food allergens include the abundance of the protein in a food, the property of short stretches of the sequence of the protein that may constitute linear IgE binding epitopes, the structural properties of the protein, its stability to heat and digestion, the food matrix the protein is in, and the antimicrobial activity to the microbial flora of the human gastrointestinal tract.
allergenicity
epitopes
vicilin leader peptide cC3C
plant allergen structure
1. Introduction
Food allergies are adverse immune responses to foods. The symptoms of a food allergy range from mild hives and itching to life-threatening anaphylaxis. In the US, it was estimated that up to 26 million adults [1] and 6 million children [2] have food allergies. Depending on the methods of studies, the sub-population suffering from food allergies in Europe was estimated between 0.8% (by positive food challenge) to 19.9% (by a survey of self-reported food allergies) [3], and the overall food allergy prevalence in Asia is comparable to that in the West [4]. Food allergies are among the top causes of anaphylaxis that lead to children’s visits to emergency departments in the United States [5]. In addition, the situation is worsening as food allergy prevalence has increased in the past few decades [2][6][7][8]. Most food allergy reactions are the immediate type that happens within hours of food intake. They are reactions to proteins (except for a small number of cases, see below) mediated by immunoglobulin E (IgE) antibodies. Food allergy reactions happen because the immune system has previously, for unknown reasons, mistaken a food protein as a dangerous invader, switched the class of T helper cells that determines whether the B cells produce IgG or IgE, and developed IgE antibodies against the protein in a so-called sensitization stage. Sensitization to food can happen in an individual when that person consumes it for the first time. It can also occur in people even though they have been eating the food safely for years to decades. In the sensitization stage, abnormal immune responses promote the class switching of B-cells to produce IgE antibodies specific to a food protein and clonal expansion of naive and IgE+ memory B-cell populations [9]. IgE molecules bind to the surface of mast cells and basophils through their association with the high-affinity IgE receptor FcεRI. Subsequent consumption of the food by the patient leads to allergen cross-linking of the IgE antibodies, which, in turn, results in the initiation of allergic reactions via signaling through the high-affinity receptor for the Fc region of immunoglobulin E (IgE) or FcεRI [10][11]. Extensive research on food allergies has been conducted in recent years. Most of these efforts involved studying the genetic, environmental, and other factors that cause a sub-population to develop a food allergy [12]. In comparison, less research has been devoted to studies on the offending allergens.
2. Food Allergens
Allergens are given names by the Allergen Nomenclature Sub-committee, which operates under the auspices of the World Health Organization (WHO) and the International Union of Immunological Societies (IUIS) [13]. The approved name contains three parts with separations by a space, three letters for the genus being the first part, one letter for the species as the second part, and an Arabic number. The letters are those at the beginning of the genus and species names and the Arabic number indicates the order of the identification of the allergens in that species [13]. The fourth letter of the genus and/or the second letter of the species is included when necessary to remove ambiguity. The WHO/IUIS Allergen Nomenclature Sub-committee also maintains a database of allergens with designated names. This database currently contains 248 food allergens of plant sources from 76 species.
In the early days of plant protein studies, they were classified based on their solubility and extractability in a series of solvent extractions and they were grouped into four groups (albumins, globulins, prolamins, and glutelins) [14]. With the advancement of knowledge of the function, biochemical, and molecular properties of proteins, plant proteins can also be classified into three groups based on their functions, structural and metabolic proteins, protective proteins, and storage proteins. Metabolic proteins can be named by their biochemical activity, while storage proteins are generally those without a known cellular activity other than the storage of nitrogen, carbon, and sulfur for the development of the next generation of the plant. Protective proteins are those with a function to defend the plant against pests, microbial pathogens, or environmental stresses. In the field of modern protein research, one of the methods of obtaining valuable information on proteins by analyzing their sequence, structure, and function is the Pfam classification of protein families based on hidden Markov model profiles [15]. At present, proteins are classified into about 19 thousand Pfam signatures [16].
The number of protein families that contain proteins from plant sources that are known to be capable of eliciting allergic responses in atopic individuals is several orders of magnitude lower compared to the total number of Pfam signatures. There are thousands of proteins in a mature plant seed [17][18], but 79% of plant food allergens belong to only 12 protein families. Table 1 listed the protein families that are known to contain more than two food allergens along with the number of known allergens in each of the families. Some of the allergens (e.g., chitinases) include two or more domains that belong to different protein families. In addition to those shown in Table 1, six protein families contain two allergens. Twenty-four allergens of plant sources belong to protein families that contain only one known allergen. Nine allergens do not belong to any of the classified Pfam families, i.e., searching the Pfam database with the allergen sequences using the online search tool at the site of the Pfam database did not find any hit. Note that minimal sequence information was available about four of the allergens in the database.
Table 1. Protein families with the most known food allergens from plant sources.
| Protein Family a | No. of Allergens | Proteins | No. of Allergens |
|---|---|---|---|
| PF00234: Tryp_alpha_amyl | 74 | Nsltp | 42 |
| 2S albumin | 26 | ||
| Others | 6 | ||
| PF00190: Cupin_1 | 39 | 11S | 19 |
| 7S | 20 | ||
| PF00235: Profilin | 27 | Profilin | |
| PF00407: Bet_v_1 | 19 | PR-10 | |
| PF01277: Oleosin | 10 | Oleosin | |
| PF00187: Chitin_bind_1 | 7 | Chitinases | |
| (PF00182: Glyco_hydro_19) | |||
| PF00314: Thaumatin | 6 | Thaumatin-like | |
| PF02704: GASA | 6 | Gibberellin regulated protein | |
| PF04702: Vicilin_N | 6 | N-terminal of vicilin cC3C antimicrobial protein | |
| PF00112: Peptidase_C1 | |||
| PF08246: Inhibitor_I29 | 3 | Cysteine protease | |
| PF00197: Kunitz_legume | 3 | Protease inhibitors | |
| PF00304: Gamma-thionin | 3 | Defensin | |
The protein family with the most food allergens from plant sources is PF00234 (the protease inhibitor/seed storage/LTP family), which has 74 known allergens. This family includes the plant nonspecific lipid transfer proteins (NsLTP), such as NsLTP1 and NsLTP2, the 2S albumin seed storage proteins, trypsin/alpha-amylase inhibitors, and other proteins. The next protein family is the Cupin-1 family, which includes 39 allergens that are in the WHO/IUIS allergen database. They are also seed storage proteins with close to half of the allergens belonging to the 11S legumins and half belonging to the 7S vicilins. Three of these allergens are now renamed as isoforms of other allergens, but their entries in the database stay due to historical reasons and literature references. With 27 food allergens, the profilin family ranked third in containing more food allergens from plant sources. Thus, the top three protein families contain more than half of the known plant food allergens. In addition, numerous other profilins are known to be pollen allergens. This indicates that the biological activities, physical–chemical properties, and conserved structures of the allergens may play a role in determining or promoting their allergenicity. The following describes the leading plant food allergen families:
Nonspecific lipid transfer proteins. Nonspecific lipid transfer proteins (NsLTPs) are found in all land plants [19]. They are small proteins with molecular masses of around ten kDa. They were demonstrated in vitro to be able to bind and transport various phospholipids to chloroplasts or mitochondria without specificity [20]. NsLTPs are plant pathogenesis-related proteins known as PR-14, and a number of them have been demonstrated to have antimicrobial activities including NsLTPs in wheat (Triticum aestivum L.) [21][22] and mung bean (Vigna radiata L. R. Wilczek) [23]. NsLTPs can be identified by an eight-cysteine residue motif (8CM). Based on the number of residues separating one cysteine from the next and the conservation of residue types at specific positions of the flanking sequences, the NsLTPs can be divided into two types. The 8CM motif for NsLTP1 is CX2VX5–7C[VLI]XY[LAV]X8–13CCXGX12DX[QKR]X2CXCX16–21PX2CX13–15C, and that for NsLTP2 is CX4LX2CX9–11P[ST]X2CCX5QX2–4C[LF]CX2[ALI]X[DN]PX10–12[KR]X4–5CX3–4PX0–2C [24], where X with a subscript number represents the number of non-conserved amino acids residues and allowed residue variation at a single position is placed in a square bracket. Thus, C1X7–10C2X12–17C3C4X8–19C5XC6X19–24C7X4–15C8 can be used to describe the 8CM of the plant NsLTPs, where the Cys residues are numbered from 1 to 8. The functions of NsLTPs are not well understood, but their expression levels are known to be high in most tissues, indicating that they may be essential for the reproduction and survival of plants. Four NsLTP2s and 38 NsLTP1s are known to be food allergens. Known NsLTP food allergens from the major allergen sources recognized by US Food and Drug Administration (FDA) include peanut (Arachis hypogaea L.) allergen Ara h 9 [25] and Ara h 17 [26], almond (Prunus dulcis (Mill.) D.A.Webb) allergen Pru du 3 [26], chestnut (Castanea sativa Mill.) allergen Cas s 8 [27], hazelnut (Corylus avellana L.) allergen Cor a 8 [28], walnut (Juglans regia L.) allergen Jug r 3 [29], and wheat allergen Tri a 14 [30]. Furthermore, the NsLTPs from many plants not used for food are known to be pollen allergens.
The crystal structure Cor a 8 [31] was the first structure reported for an NsLTP food allergen from the major allergen sources, though that of wheat allergen Tri a 14 [32] and the solution structure of Tri a 14 [33] were reported many years ago before it was identified as a food allergen. As shown in Figure 1A, the cysteines in the 8CM of Cor a 8 form four disulfide bonds. Protein structures were generated with the CCP4MG program [34]. The structures of many other NsLTP1 food and pollen allergens are also available. The conservation of these disulfide bond connectivities (between C1–C6, C2–C3, C4–C7, and C5–C8) in NsLTP1s maintains the tertiary contacts of the secondary structural elements and ensures a stable hydrophobic cavity for lipid binding [35]. Moreover, the structure of rice (Oryza sativa L.) NsLTP2 was determined by NMR, which showed an overall structure similar to that of an NsLTP1. The disulfide bond connectivities in NsLTP2 (C1–C5, C2–C3, C4–C7, and C6–C8) are different from those in NsLTP1 [36].

Figure 1. Structures of representative members of protein families that contain the majority of the known food allergens of plant origin. The name of the allergen or protein and the protein family/subfamily is indicated below every structure following the (A–L) sequence label of the individual panels. The coordinates of the structures were downloaded from the WorldWide Protein Data Bank, and the graphics displays were generated using the CCP4MG program. The PDB codes for the structures are included in the figure labels along with the names of the allergens. Each structure is shown as a ribbon diagram with a blend-through coloring scheme displaying the N-terminal blue and the C-terminal red, except for the multimeric allergens Ara h 1 and Ara h 3 where the monomers were blended through different color ranges. Two panels of Ara h 1 are presented, with the right panel being the left panel rotated about a horizontal axis parallel to the paper pointing to the right. The side chains of cysteines that are involved in disulfide bonds are shown as ball-and-stick. Cysteines that are conserved in well-defined sequence motifs are labeled with their numbering in the motifs (see text).
2S albumins. Plant proteins coagulable by heat and soluble in water were called albumins in the early 20th century for their properties that resembled hen egg albumin [14]. The 2S albumins migrated with a 2S sedimentation coefficient during sucrose gradient centrifugation [37]. The 2S albumins also contain an 8CM similar to that of the NsLTPs but with longer sequences separating C2 and C3 and C6 and C7. Seed storage proteins are believed to accumulate in developing seeds to act as a nitrogen reserve for germination [38][39]. The 2S albumins were considered to be a major group of storage proteins in many dicotyledonous plant species [40] that also play a role in providing sulfur reserve in the seed [37]. The 2S albumins have also been suggested to have antimicrobial activities [41][42][43]. Known 2S food allergens from the major allergen sources recognized by FDA include peanut allergen Ara h 2 [44], soybean (Glycine max) allergen Gly m 8 [45], Brazil nut (Bertholletia excelsa Silva Manso) allergen Ber e 1 [46], cashew (Anacardium occidentale L.) allergen Ana o 3 [47], hazelnut allergen Cor a 14 [48], pecan (Carya illinoinensis (Wangenh.) K.Koch) allergen Car i 1 [49], pistachio (Pistacia vera L.) allergen Pis v 1 [50], Stone pine (Pinus pinea L.) allergen Pin p 1 [51], sesame (Sesamum indicum L.) allergens Ses i 1 [52] and Ses i 2 [53], and Black walnut (Juglans nigra L.) allergens Jug n 1 and walnut allergen Jug r 1 [54].
The structures of many 2S albumins including food allergens in castor beans (Ricinus communis L.) (Ric c 1) [55], rapeseed (Brassica napus L.) (Bra n 1) [56], and Brazil nuts (Ber e 1) [57] have been reported. The first structure reported for a 2S albumin allergen from the major allergen sources is that of Ara h 6, which was determined by NMR using recombinantly expressed Ara h 6 with uniform 15N and 13C labeling [58]. Three peanut 2S albumins have been identified as food allergens. They are Ara h 2, Ara h 6, and Ara h 7. The structure of Ara h 2 was also reported (Figure 1B). It was determined by X-ray crystallography using recombinantly expressed Ara h 2 with a maltose-binding protein fused to the N-terminal to enhance its solubility and aid its crystallization [59]. These 2S albumins have the same disulfide bond connections as NsLTP2. However, Ara h 6 has an additional disulfide bond, which is formed by an extra cysteine between C6 and C7 (C6′) and another cysteine residue after C8 (C8′), as shown by one of the models of its structures determined by NMR (Figure 1C).
11S legumins. Both the 11S and the 7S seed storage proteins belong to the cupin superfamily, which was initially recognized based on a 50% sequence identity between the wheat protein germin and a slime mold (Physarum polycephalum) protein spherulin [60]. Germin is an unusually thermostable protein produced during the early phase of germination in wheat embryos. The sequence similarity was then extended to a group of germin-like proteins and globulin storage proteins. Globulins were classified as those soluble in dilute salt solution but insoluble in water [14]. After structural information on canavalin [61] and phaseolin [62] became available, sequence alignment revealed a much larger group of proteins in this superfamily, and the family was given the name cupin [63] (from the Latin term ‘cupa’ which means small barrel). The cupin superfamily contains monocupin, bicupins, and multicupins. It is known to be one of the most functionally diverse protein superfamilies [64] including various proteins with enzymatic functions, non-enzymatic transcription factors, and the 11S and 7S seed storage proteins.
The signature of the cupins includes two sequence motifs separated by an inter-motif sequence with variable length (from 11 amino acids to over a hundred residues). The first motif was defined as GX5HXHX3–4EX6G, and the second motif was characterized as GX5PXGX2HX3N. The two histidines and the glutamate in motif 1 and the histidine in motif 2 may act as ligands to bind metal ions. In many cupins with enzymatic activity, a metal ion is part of the active site [65]. However, the motifs are now known to tolerate variations and not all cupins have a metal ligand [64]. Nevertheless, residues other than those specified above can also provide metal ligand coordination [66].
The 11S globulins are the most widespread among seed storage protein groups. They are present in monocot and eudicot seeds, as well as in conifers and other gymnosperms. They are particularly abundant in legume seeds and are often called legumins. Typical legumins have molecular weights (MW) of about 300–450 kD and consist of six subunits of about 60 kD. These subunits are the products of a multigene family. Each subunit is post-translationally processed to give rise to an acidic (MW about 40 kD) chain and a basic (~20 kD) chain [67]. The acidic and the basic chains are linked by a single disulfide bond. The 11S globulins are rarely, if ever, glycosylated. This family of proteins accounts for many of the known major food allergens from the FDA-recognized major allergen sources including peanut allergen Ara h 3 [68], soybean allergen Gly m 6 [69], almond allergen Pru du 6 [70], Brazil nut allergen Ber e 2, cashew allergen Ana o 2 [71], hazelnut allergen Cor a 9 [72], macadamia (Macadamia integrifolia Maiden and Betche) allergen Mac i 2 [73][74], pecan allergen Car i 4 [75], pistachio allergen Pis v 2 [50], sesame allergens Ses i 6 and Ses i 7 [76], and walnut allergens Jug n 4 [77] and Jug r 4 [78].
The first structure reported for a legumin food allergen from the FDA-recognized major food sources is that of Gly m 6, which was determined before it was designated as a food allergen [13][79]. The 11S seed storage proteins in many species have more than one type of subunit and five for Gly m 6 [79]. Mature 11S proteins are hexamers that can be composed of different subunits, making it problematic for crystallization. The crystal structure of Gly m 6 was determined by purifying the protein from genetically modified soybeans with four of the subunits of the 11S protein deleted. The peanut 11S food allergen is also known to be coded by at least five different genes [80]. Nevertheless, the population of the mature protein that is composed of the translation from a single gene may be high, and the crystallization of Ara h 3 from wild-type peanuts was successful [81]. The first crystal structure of a peanut allergen was that of Ara h 3 (Figure 1D) [82]. The structure of another 11S allergen from the FDA-recognized major food sources, Pru du 6, was also determined [83]. Generally, the 11S allergens are a dimer of trimers. While the doughnut-shaped trimer was made up of three subunits by head-to-tail associations, the back-to-back binding of the trimers forms the hexameric structure of the native molecule. The dimerization of the trimers buries the N-terminus of the basic subdomain which was generated as a result of the cleavage at a conserved peptidase recognition site. The C-terminal of the acidic domain, however, moved away before the trimer–trimer interface to facilitate the packing of the mature hexamer, making it impossible to express the 11S allergen recombinantly with most of the commonly used strategies [83]. The structure of an 11S putative allergens purified from coconut was also determined recently [84][85].
7S vicilins. The 7S globulins are called vicilins, and they are also present in flowering plants and other spermatophytes. Vicilins are trimeric proteins of MW of ~150–190 kD, with a typical subunit MW of ~50 kD. No disulfide bond was found in vicilins, but proteolytic processing and glycosylation may occur [86][87]. Thus, the subunit structure of vicilins revealed by SDS-PAGE is similar in the absence or presence of reducing agents. Vicilins also account for many known major food allergens from the FDA-recognized major allergen sources including peanut allergen Ara h 1 [88], soybean allergen Gly m 5 [69], almond vicilin [89], cashew allergen Ana o 1 [90], hazelnut allergen Cor a 11 and Cor a 16 [91][92], macadamia allergen Mac i 1 [73], pecan allergen Car i 2 [93], pistachio allergen Pis v 3 [94], Korean pine (Pinus koraiensis Siebold and Zucc.) allergen Pin k 2 [95][96], walnut allergens Jug n 2 and Jug r 2 [97], and sesame allergen Ses i 3 [53].
Vicilin leader peptides. Vicilins from some species, such as pea (Pisum sativum L.) allergen Pis s 1 [98], contain just the di-cupin region and a signal peptide. However, vicilins from other species are known to have a variable region between the signal peptide, which can be predicted [99], and the C-terminal di-cupin domains [100][101][102]. This variable leader peptide (VLP) was also called vicilin leader peptide. When they are found in a food independent of the mature vicilin protein with demonstrated allergenicity, they are designated as vicilin iso allergens by the WHO/IUUS Allergen Nomenclature Subcommittee, e.g., Ara h 1.0101 (26–84). In many vicilins, this region can consist of one (as in almond vicilin allergen [89] and peanut allergen Ara h 1 [88]) or more (as in pecan allergen Car i 2 [93]) repeats of a coupled-C3C (cC3C) motif which has a quintet of cysteines arranged as a pair of CX3C linked by 8–12 amino acids. Interestingly, none of this variable region, part of it, or the entirety of the region can be found in the native vicilin purified from the seeds, depending on the plant species. The cC3C area of macadamia nut vicilin was reported to have antimicrobial activities [103]. Available data on the cC3C repeat in the variable region of vicilin food allergens are summarized in Figure 2.
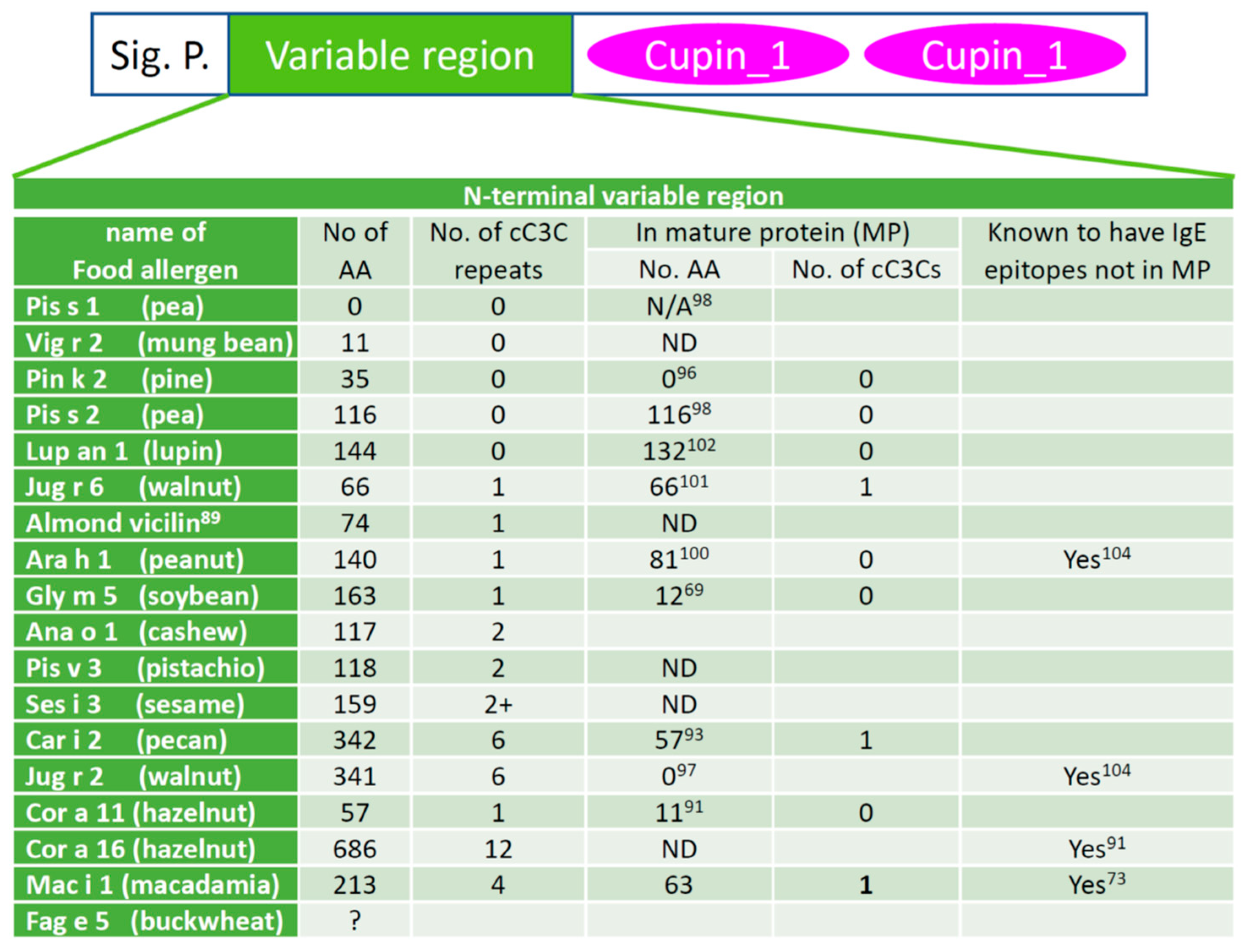
Figure 2. cC3C repeats of the variable leader peptide of known vicilin allergens. The domain structure of vicilin is shown at the top. Several features of the N-terminal variable region of vicilin between the signal peptide (Sig. P.) and the C-terminal cupin domains are shown below the domain structure. A question mark in the second column indicates the full sequence of the variable region of the allergen is not available. “ND” means no data available, and superscripts indicate the reference number of the cited literature. The variable leader peptides are derived by determining the signal peptides and the N-terminal peptides of the natural allergens. The sequences of the allergens were downloaded from the protein database at NCBI. The signal peptides were predicted using SignalP 6.0 and the N-termini were those reported in the relevant references. The reference numbers are given as superscripts in the table cells. Almond vicilin was reported to be an allergen but does not have an Allergen Nomenclature Subcommittee designated allergen name.
References
- Sicherer, S.H.; Sampson, H.A. Food allergy: A review and update on epidemiology, pathogenesis, diagnosis, prevention, and management. J. Allergy Clin. Immunol. 2018, 141, 41–58.
- Gupta, R.S.; Springston, E.E.; Warrier, M.R.; Smith, B.; Kumar, R.; Pongracic, J.; Holl, J.L. The prevalence, severity, and distribution of childhood food allergy in the United States. Pediatrics 2011, 128, e9–e17.
- Spolidoro, G.C.I.; Amera, Y.T.; Ali, M.M.; Nyassi, S.; Lisik, D.; Ioannidou, A.; Rovner, G.; Khaleva, E.; Venter, C.; van Ree, R.; et al. FREQUENCY of food allergy in Europe: An updated systematic review and meta-analysis. Allergy 2023, 78, 351–368.
- Lee, A.J.; Thalayasingam, M.; Lee, B.W. Food allergy in Asia: How does it compare? Asia Pac. Allergy 2013, 3, 3–14.
- Motosue, M.S.; Bellolio, M.F.; Van Houten, H.K.; Shah, N.D.; Campbell, R.L. National trends in emergency department visits and hospitalizations for food-induced anaphylaxis in US children. Pediatr. Allergy Immunol. 2018, 29, 538–544.
- Gupta, R.S.; Warren, C.M.; Smith, B.M.; Jiang, J.L.; Blumenstock, J.A.; Davis, M.M.; Schleimer, R.P.; Nadeau, K.C. Prevalence and severity of food allergies among US adults. JAMA Netw. Open 2019, 2, e185630.
- Sicherer, S.H.; Munoz-Furlong, A.; Sampson, H.A. Prevalence of peanut and tree nut allergy in the United States determined by means of a random digit dial telephone survey: A 5-year follow-up study. J. Allergy Clin. Immunol. 2003, 112, 1203–1207.
- Kumfer, A.M.; Commins, S.P. Primary prevention of food allergy. Curr. Allergy Asthma Rep. 2019, 19, 7.
- Bhalla, P.L.; Singh, M.B. Biotechnology-based allergy diagnosis and vaccination. Trends Biotechnol. 2008, 26, 153–161.
- Galli, S.J.; Tsai, M.; Piliponsky, A.M. The development of allergic inflammation. Nature 2008, 454, 445–454.
- Fattakhova, G.; Masilamani, M.; Borrego, F.; Gilfillan, A.M.; Metcalfe, D.D.; Coligan, J.E. The high-affinity immunoglobulin-E receptor (FcepsilonRI) is endocytosed by an AP-2/clathrin-independent, dynamin-dependent mechanism. Traffic 2006, 7, 673–685.
- Krempski, J.W.; Dant, C.; Nadeau, K.C. The origins of allergy from a systems approach. Ann. Allergy Asthma Immunol. 2020, 125, 507–516.
- IUIS/WHO-Allergen-Nomenclature-Subcommittee. Allergen nomenclature. Bull. World Health Organ. 1994, 72, 797–806.
- Osborne, T.B. The Vegetable Proteins, 2nd ed.; Green and Co.: London, UK, 1924; p. 440.
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432.
- Paysan-Lafosse, T.; Blum, M.; Chuguransky, S.; Grego, T.; Pinto, B.L.; Salazar, G.A.; Bileschi, M.L.; Bork, P.; Bridge, A.; Colwell, L.; et al. InterPro in 2022. Nucleic Acids Res. 2023, 51, D418–D427.
- Gallardo, K.; Job, C.; Groot, S.P.; Puype, M.; Demol, H.; Vandekerckhove, J.; Job, D. Proteomic analysis of arabidopsis seed germination and priming. Plant Physiol. 2001, 126, 835–848.
- Mooney, B.P.; Krishnan, H.B.; Thelen, J.J. High-throughput peptide mass fingerprinting of soybean seed proteins: Automated workflow and utility of UniGene expressed sequence tag databases for protein identification. Phytochemistry 2004, 65, 1733–1744.
- Salminen, T.A.; Blomqvist, K.; Edqvist, J. Lipid transfer proteins: Classification, nomenclature, structure, and function. Planta 2016, 244, 971–997.
- Kader, J.C.; Julienne, M.; Vergnolle, C. Purification and characterization of a spinach-leaf protein capable of transferring phospholipids from liposomes to mitochondria or chloroplasts. Eur. J Biochem. 1984, 139, 411–416.
- Sun, J.Y.; Gaudet, D.A.; Lu, Z.X.; Frick, M.; Puchalski, B.; Laroche, A. Characterization and antifungal properties of wheat nonspecific lipid transfer proteins. Mol. Plant Microbe Interact. 2008, 21, 346–360.
- Odintsova, T.I.; Slezina, M.P.; Istomina, E.A.; Korostyleva, T.V.; Kovtun, A.S.; Kasianov, A.S.; Shcherbakova, L.A.; Kudryavtsev, A.M. Non-specific lipid transfer proteins in Triticum kiharae Dorof. et Migush.: Identification, characterization and expression profiling in response to pathogens and resistance inducers. Pathogens 2019, 8, 221.
- Wang, S.Y.; Wu, J.H.; Ng, T.B.; Ye, X.Y.; Rao, P.F. A non-specific lipid transfer protein with antifungal and antibacterial activities from the mung bean. Peptides 2004, 25, 1235–1242.
- Wang, N.J.; Lee, C.C.; Cheng, C.S.; Lo, W.C.; Yang, Y.F.; Chen, M.N.; Lyu, P.C. Construction and analysis of a plant non-specific lipid transfer protein database (nsLTPDB). BMC Genom. 2012, 13 (Suppl. S1), S9.
- Krause, S.; Reese, G.; Randow, S.; Zennaro, D.; Quaratino, D.; Palazzo, P.; Ciardiello, M.A.; Petersen, A.; Becker, W.M.; Mari, A. Lipid transfer protein (Ara h 9) as a new peanut allergen relevant for a Mediterranean allergic population. J. Allergy Clin. Immunol. 2009, 124, 771–778.e775.
- WHO/IUIS-Allergen-Nomenclature-Sub-Committee. Allergen Nomenclature. Available online: http://www.allergen.org/ (accessed on 14 April 2023).
- Sanchez-Monge, R.; Blanco, C.; Lopez-Torrejon, G.; Cumplido, J.; Recas, M.; Figueroa, J.; Carrillo, T.; Salcedo, G. Differential allergen sensitization patterns in chestnut allergy with or without associated latex-fruit syndrome. J. Allergy Clin. Immunol. 2006, 118, 705–710.
- Pastorello, E.A.; Vieths, S.; Pravettoni, V.; Farioli, L.; Trambaioli, C.; Fortunato, D.; Luttkopf, D.; Calamari, M.; Ansaloni, R.; Scibilia, J.; et al. Identification of hazelnut major allergens in sensitive patients with positive double-blind, placebo-controlled food challenge results. J. Allergy Clin. Immunol. 2002, 109, 563–570.
- Pastorello, E.A.; Farioli, L.; Pravettoni, V.; Robino, A.M.; Scibilia, J.; Fortunato, D.; Conti, A.; Borgonovo, L.; Bengtsson, A.; Ortolani, C. Lipid transfer protein and vicilin are important walnut allergens in patients not allergic to pollen. J. Allergy Clin. Immunol. 2004, 114, 908–914.
- Sander, I.; Rozynek, P.; Rihs, H.P.; van Kampen, V.; Chew, F.T.; Lee, W.S.; Kotschy-Lang, N.; Merget, R.; Bruning, T.; Raulf-Heimsoth, M. Multiple wheat flour allergens and cross-reactive carbohydrate determinants bind IgE in baker’s asthma. Allergy 2011, 66, 1208–1215.
- Offermann, L.R.; Bublin, M.; Perdue, M.L.; Pfeifer, S.; Dubiela, P.; Borowski, T.; Chruszcz, M.; Hoffmann-Sommergruber, K. Structural and functional characterization of the hazelnut allergen Cor a 8. J. Agric. Food Chem. 2015, 63, 9150–9158.
- Charvolin, D.; Douliez, J.P.; Marion, D.; Cohen-Addad, C.; Pebay-Peyroula, E. The crystal structure of a wheat nonspecific lipid transfer protein (ns-LTP1) complexed with two molecules of phospholipid at 2.1 A resolution. Eur. J. Biochem. 1999, 264, 562–568.
- Tassin-Moindrot, S.; Caille, A.; Douliez, J.P.; Marion, D.; Vovelle, F. The wide binding properties of a wheat nonspecific lipid transfer protein. Solution structure of a complex with prostaglandin B2. Eur. J. Biochem. 2000, 267, 1117–1124.
- McNicholas, S.; Potterton, E.; Wilson, K.S.; Noble, M.E. Presenting your structures: The CCP4mg molecular-graphics software. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 386–394.
- Sy, D.; Le Gravier, Y.; Goodfellow, J.; Vovelle, F. Protein stability and plasticity of the hydrophobic cavity in wheat ns-LTP. J. Biomol. Struct. Dyn. 2003, 21, 15–29.
- Samuel, D.; Liu, Y.J.; Cheng, C.S.; Lyu, P.C. Solution structure of plant nonspecific lipid transfer protein-2 from rice (Oryza sativa). J. Biol. Chem. 2002, 277, 35267–35273.
- Youle, R.J.; Huang, A.H.C. Occurrence of low-molecular weight and high cysteine containing albumin storage proteins in oilseeds of diverse species. Am. J. Bot. 1981, 68, 44–48.
- Utsumi, S. Plant food protein engineering. Adv. Food Nutr. Res. 1992, 36, 89–208.
- Shewry, P.R. Plant storage proteins. Biol. Rev. Camb. Philos. Soc. 1995, 70, 375–426.
- Shewry, P.R.; Napier, J.A.; Tatham, A.S. Seed storage proteins: Structures and biosynthesis. Plant Cell 1995, 7, 945–956.
- Candido Ede, S.; Pinto, M.F.; Pelegrini, P.B.; Lima, T.B.; Silva, O.N.; Pogue, R.; Grossi-de-Sa, M.F.; Franco, O.L. Plant storage proteins with antimicrobial activity: Novel insights into plant defense mechanisms. FASEB J. 2011, 25, 3290–3305.
- Odintsova, T.I.; Rogozhin, E.A.; Sklyar, I.V.; Musolyamov, A.K.; Kudryavtsev, A.M.; Pukhalsky, V.A.; Smirnov, A.N.; Grishin, E.V.; Egorov, T.A. Antifungal activity of storage 2S albumins from seeds of the invasive weed dandelion Taraxacum officinale Wigg. Protein Pept. Lett. 2010, 17, 522–529.
- Pelegrini, P.B.; Noronha, E.F.; Muniz, M.A.; Vasconcelos, I.M.; Chiarello, M.D.; Oliveira, J.T.; Franco, O.L. An antifungal peptide from passion fruit (Passiflora edulis) seeds with similarities to 2S albumin proteins. Biochim. Biophys. Acta 2006, 1764, 1141–1146.
- Kleber-Janke, T.; Crameri, R.; Appenzeller, U.; Schlaak, M.; Becker, W.M. Selective cloning of peanut allergens, including profilin and 2S albumins, by phage display technology. Int. Arch. Allergy Immunol. 1999, 119, 265–274.
- Klemans, R.J.; Knol, E.F.; Michelsen-Huisman, A.; Pasmans, S.G.; de Kruijf-Broekman, W.; Bruijnzeel-Koomen, C.A.; van Hoffen, E.; Knulst, A.C. Components in soy allergy diagnostics: Gly m 2S albumin has the best diagnostic value in adults. Allergy 2013, 68, 1396–1402.
- Pastorello, E.A.; Farioli, L.; Pravettoni, V.; Ispano, M.; Conti, A.; Ansaloni, R.; Rotondo, F.; Incorvaia, C.; Bengtsson, A.; Rivolta, F.; et al. Sensitization to the major allergen of Brazil nut is correlated with the clinical expression of allergy. J. Allergy Clin. Immunol. 1998, 102, 1021–1027.
- Robotham, J.M.; Wang, F.; Seamon, V.; Teuber, S.S.; Sathe, S.K.; Sampson, H.A.; Beyer, K.; Seavy, M.; Roux, K.H. Ana o 3, an important cashew nut (Anacardium occidentale L.) allergen of the 2S albumin family. J. Allergy Clin. Immunol. 2005, 115, 1284–1290.
- Garino, C.; Zuidmeer, L.; Marsh, J.; Lovegrove, A.; Morati, M.; Versteeg, S.; Schilte, P.; Shewry, P.; Arlorio, M.; van Ree, R. Isolation, cloning, and characterization of the 2S albumin: A new allergen from hazelnut. Mol. Nutr. Food Res. 2010, 54, 1257–1265.
- Sharma, G.M.; Irsigler, A.; Dhanarajan, P.; Ayuso, R.; Bardina, L.; Sampson, H.A.; Roux, K.H.; Sathe, S.K. Cloning and characterization of 2S albumin, Car i 1, a major allergen in pecan. J. Agric. Food Chem. 2011, 59, 4130–4139.
- Ahn, K.; Bardina, L.; Grishina, G.; Beyer, K.; Sampson, H.A. Identification of two pistachio allergens, Pis v 1 and Pis v 2, belonging to the 2S albumin and 11S globulin family. Clin. Exp. Allergy 2009, 39, 926–934.
- Cabanillas, B.; Crespo, J.F.; Maleki, S.J.; Rodriguez, J.; Novak, N. Pin p 1 is a major allergen in pine nut and the first food allergen described in the plant group of gymnosperms. Food Chem. 2016, 210, 70–77.
- Pastorello, E.A.; Varin, E.; Farioli, L.; Pravettoni, V.; Ortolani, C.; Trambaioli, C.; Fortunato, D.; Giuffrida, M.G.; Rivolta, F.; Robino, A.; et al. The major allergen of sesame seeds (Sesamum indicum) is a 2S albumin. J. Chromatogr. B Biomed. Sci. Appl. 2001, 756, 85–93.
- Beyer, K.; Bardina, L.; Grishina, G.; Sampson, H.A. Identification of sesame seed allergens by 2-dimensional proteomics and Edman sequencing: Seed storage proteins as common food allergens. J. Allergy Clin. Immunol. 2002, 110, 154–159.
- Teuber, S.S.; Dandekar, A.M.; Peterson, W.R.; Sellers, C.L. Cloning and sequencing of a gene encoding a 2S albumin seed storage protein precursor from English walnut (Juglans regia), a major food allergen. J. Allergy Clin. Immunol. 1998, 101, 807–814.
- Pantoja-Uceda, D.; Bruix, M.; Gimenez-Gallego, G.; Rico, M.; Santoro, J. Solution structure of RicC3, a 2S albumin storage protein from Ricinus communis. Biochemistry 2003, 42, 13839–13847.
- Pantoja-Uceda, D.; Palomares, O.; Bruix, M.; Villalba, M.; Rodriguez, R.; Rico, M.; Santoro, J. Solution structure and stability against digestion of rproBnIb, a recombinant 2S albumin from rapeseed: Relationship to its allergenic properties. Biochemistry 2004, 43, 16036–16045.
- Rundqvist, L.; Tengel, T.; Zdunek, J.; Bjorn, E.; Schleucher, J.; Alcocer, M.J.; Larsson, G. Solution structure, copper binding and backbone dynamics of recombinant Ber e 1-the major allergen from Brazil nut. PLoS ONE 2012, 7, e46435.
- Lehmann, K.; Schweimer, K.; Reese, G.; Randow, S.; Suhr, M.; Becker, W.M.; Vieths, S.; Rosch, P. Structure and stability of 2S albumin-type peanut allergens: Implications for the severity of peanut allergic reactions. Biochem. J. 2006, 395, 463–472.
- Mueller, G.A.; Gosavi, R.A.; Pomes, A.; Wunschmann, S.; Moon, A.F.; London, R.E.; Pedersen, L.C. Ara h 2: Crystal structure and IgE binding distinguish two subpopulations of peanut allergic patients by epitope diversity. Allergy 2011, 66, 878–885.
- Lane, B.G.; Bernier, F.; Dratewka-Kos, E.; Shafai, R.; Kennedy, T.D.; Pyne, C.; Munro, J.R.; Vaughan, T.; Walters, D.; Altomare, F. Homologies between members of the germin gene family in hexaploid wheat and similarities between these wheat germins and certain Physarum spherulins. J. Biol. Chem. 1991, 266, 10461–10469.
- Ko, T.P.; Ng, J.D.; McPherson, A. The three-dimensional structure of canavalin from jack bean (Canavalia ensiformis). Plant Physiol. 1993, 101, 729–744.
- Lawrence, M.C.; Izard, T.; Beuchat, M.; Blagrove, R.J.; Colman, P.M. Structure of phaseolin at 2.2 A resolution. Implications for a common vicilin/legumin structure and the genetic engineering of seed storage proteins. J. Mol. Biol. 1994, 238, 748–776.
- Dunwell, J.M. Cupins: A new superfamily of functionally diverse proteins that include germins and plant storage proteins. Biotechnol. Genet. Eng. Rev. 1998, 15, 1–32.
- Dunwell, J.M.; Purvis, A.; Khuri, S. Cupins: The most functionally diverse protein superfamily? Phytochemistry 2004, 65, 7–17.
- Hansen, T.; Schlichting, B.; Felgendreher, M.; Schonheit, P. Cupin-type phosphoglucose isomerases (Cupin-PGIs) constitute a novel metal-dependent PGI family representing a convergent line of PGI evolution. J. Bacteriol. 2005, 187, 1621–1631.
- Jin, T.; Wang, Y.; Chen, Y.W.; Fu, T.J.; Kothary, M.H.; McHugh, T.H.; Zhang, Y. Crystal structure of the Korean pine (Pinus koraiensis) 7S seed storage protein with copper ligands. J. Agric. Food Chem. 2014, 62, 222–228.
- Mills, E.N.; Jenkins, J.; Marigheto, N.; Belton, P.S.; Gunning, A.P.; Morris, V.J. Allergens of the cupin superfamily. Biochem. Soc. Trans. 2002, 30, 925–929.
- Rabjohn, P.; Helm, E.M.; Stanley, J.S.; West, C.M.; Sampson, H.A.; Burks, A.W.; Bannon, G.A. Molecular cloning and epitope analysis of the peanut allergen Ara h 3. J. Clin. Investig. 1999, 103, 535–542.
- Holzhauser, T.; Wackermann, O.; Ballmer-Weber, B.K.; Bindslev-Jensen, C.; Scibilia, J.; Perono-Garoffo, L.; Utsumi, S.; Poulsen, L.K.; Vieths, S. Soybean (Glycine max) allergy in Europe: Gly m 5 (beta-conglycinin) and Gly m 6 (glycinin) are potential diagnostic markers for severe allergic reactions to soy. J. Allergy Clin. Immunol. 2009, 123, 452–458.
- Willison, L.N.; Tripathi, P.; Sharma, G.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Cloning, expression and patient IgE reactivity of recombinant Pru du 6, an 11S globulin from almond. Int. Arch. Allergy Immunol. 2011, 156, 267–281.
- Wang, F.; Robotham, J.M.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Ana o 2, a major cashew (Anacardium occidentale L.) nut allergen of the legumin family. Int. Arch. Allergy Immunol. 2003, 132, 27–39.
- Beyer, K.; Grishina, G.; Bardina, L.; Grishin, A.; Sampson, H.A. Identification of an 11S globulin as a major hazelnut food allergen in hazelnut-induced systemic reactions. J. Allergy Clin. Immunol. 2002, 110, 517–523.
- Kabasser, S.; Pratap, K.; Kamath, S.; Taki, A.C.; Dang, T.; Koplin, J.; Perrett, K.; Hummel, K.; Radauer, C.; Breiteneder, H.; et al. Identification of vicilin, legumin and antimicrobial peptide 2a as macadamia nut allergens. Food Chem. 2022, 370, 131028.
- Zhang, Y.; Bhardwaj, S.R.; Lyu, S.C.; Chinthrajah, S.; Nadeau, K.C.; Li, C. Expression, purification, characterization, and patient IgE reactivity of new macadamia nut iso-allergen. Protein Expr. Purif. 2022, 203, 106211.
- Sharma, G.M.; Irsigler, A.; Dhanarajan, P.; Ayuso, R.; Bardina, L.; Sampson, H.A.; Roux, K.H.; Sathe, S.K. Cloning and characterization of an 11S legumin, Car i 4, a major allergen in pecan. J. Agric. Food Chem. 2011, 59, 9542–9552.
- Beyer, K.; Grishina, G.; Bardina, L.; Sampson, H.A. Identification of 2 new sesame seed allergens: Ses i 6 and Ses i 7. J. Allergy Clin. Immunol. 2007, 119, 1554–1556.
- Zhang, Y.Z.; Du, W.X.; Fan, Y.; Yi, J.; Lyu, S.C.; Nadeau, K.C.; Thomas, A.L.; McHugh, T. Purification and characterization of a black walnut (Juglans nigra) allergen, Jug n 4. J. Agric. Food Chem. 2017, 65, 454–462.
- Wallowitz, M.; Peterson, W.R.; Uratsu, S.; Comstock, S.S.; Dandekar, A.M.; Teuber, S.S. Jug r 4, a legumin group food allergen from walnut (Juglans regia Cv. Chandler). J. Agric. Food Chem. 2006, 54, 8369–8375.
- Adachi, M.; Kanamori, J.; Masuda, T.; Yagasaki, K.; Kitamura, K.; Mikami, B.; Utsumi, S. Crystal structure of soybean 11S globulin: Glycinin A3B4 homohexamer. Proc. Natl. Acad. Sci. USA 2003, 100, 7395–7400.
- Yan, Y.-S.; Lin, X.-D.; Zhang, Y.-S.; Wang, L.; Wu, K.; Huang, S.-Z. Isolation of peanut genes encoding arachins and conglutins by expressed sequence tags. Plant Sci. 2005, 169, 439–445.
- Jin, T.; Howard, A.; Zhang, Y.Z. Purification, crystallization and initial crystallographic characterization of peanut major allergen Ara h 3. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007, 63, 848–851.
- Jin, T.; Guo, F.; Chen, Y.-W.; Howard, A.; Zhang, Y.-Z. Crystal structure of Ara h 3, a major allergen in peanut. Mol. Immunol. 2009, 46, 1796–1804.
- Jin, T.; Albillos, S.M.; Guo, F.; Howard, A.; Fu, T.J.; Kothary, M.H.; Zhang, Y.Z. Crystal structure of prunin-1, a major component of the almond (Prunus dulcis) allergen amandin. J. Agric. Food Chem. 2009, 57, 8643–8651.
- Vajravijayan, S.; Nandhagopal, N.; Gunasekaran, K. Crystal structure determination and analysis of 11S coconut allergen: Cocosin. Mol. Immunol. 2017, 92, 132–135.
- Jin, T.; Wang, C.; Zhang, C.; Wang, Y.; Chen, Y.W.; Guo, F.; Howard, A.; Cao, M.J.; Fu, T.J.; McHugh, T.H.; et al. Crystal structure of cocosin, a potential food allergen from coconut (Cocos nucifera). J. Agric. Food Chem. 2017, 65, 7560–7568.
- Gatehouse, J.A.; Lycett, G.W.; Croy, R.R.; Boulter, D. The post-translational proteolysis of the subunits of vicilin from pea (Pisum sativum L.). Biochem. J. 1982, 207, 629–632.
- Lopez-Torrejon, G.; Salcedo, G.; Martin-Esteban, M.; Diaz-Perales, A.; Pascual, C.Y.; Sanchez-Monge, R. Len c 1, a major allergen and vicilin from lentil seeds: Protein isolation and cDNA cloning. J. Allergy Clin. Immunol. 2003, 112, 1208–1215.
- Burks, A.W.; Cockrell, G.; Stanley, J.S.; Helm, R.M.; Bannon, G.A. Recombinant peanut allergen Ara h I expression and IgE binding in patients with peanut hypersensitivity. J. Clin. Investig. 1995, 96, 1715–1721.
- Che, H.L.; Zhang, Y.Z.; Lyu, S.C.; Nadeau, K.C.; McHugh, T. Identification of almond (Prunus dulcis) vicilin as a food allergen. J. Agric. Food Chem. 2019, 67, 425–432.
- Wang, F.; Robotham, J.M.; Teuber, S.S.; Tawde, P.; Sathe, S.K.; Roux, K.H. Ana o 1, a cashew (Anacardium occidental) allergen of the vicilin seed storage protein family. J. Allergy Clin. Immunol. 2002, 110, 160–166.
- Lauer, I.; Foetisch, K.; Kolarich, D.; Ballmer-Weber, B.K.; Conti, A.; Altmann, F.; Vieths, S.; Scheurer, S. Hazelnut (Corylus avellana) vicilin Cor a 11: Molecular characterization of a glycoprotein and its allergenic activity. Biochem. J. 2004, 383, 327–334.
- Mattsson, L.; Holmqvist, M.; Porsch, H.; Larsson, H.; Pontoppidan, B.; Valcour, A.; Lidholm, J. A new vicilin-like allergen in hazelnut giving rise to a spectrum of IgE-binding low-molecular-weight N-terminal fragments. Clin. Exp. Allergy 2022, 52, 1208–1212.
- Zhang, Y.; Lee, B.; Du, W.X.; Lyu, S.C.; Nadeau, K.C.; Grauke, L.J.; Zhang, Y.; Wang, S.; Fan, Y.; Yi, J.; et al. Identification and characterization of a new pecan allergen, Car i 2. J. Agric. Food Chem. 2016, 64, 4146–4151.
- Willison, L.N.; Tawde, P.; Robotham, J.M.; Penney, R.M.t.; Teuber, S.S.; Sathe, S.K.; Roux, K.H. Pistachio vicilin, Pis v 3, is immunoglobulin E-reactive and cross-reacts with the homologous cashew allergen, Ana o 1. Clin. Exp. Allergy 2008, 38, 1229–1238.
- Zhang, Y.Z.; Du, W.X.; Fan, Y.T.; Yi, J.; Lyu, S.C.; Nadeau, K.C.; McHugh, T.H. Identification, characterization, and initial epitope mapping of pine nut allergen Pin k 2. Food Res. Int. 2016, 90, 268–274.
- Jin, T.; Albillos, S.M.; Chen, Y.W.; Kothary, M.H.; Fu, T.J.; Zhang, Y.Z. Purification and characterization of the 7S vicilin from Korean pine (Pinus koraiensis). J. Agric. Food Chem. 2008, 56, 8159–8165.
- Teuber, S.S.; Jarvis, K.C.; Dandekar, A.M.; Peterson, W.R.; Ansari, A.A. Identification and cloning of a complementary DNA encoding a vicilin-like proprotein, jug r 2, from english walnut kernel (Juglans regia), a major food allergen. J. Allergy Clin. Immunol. 1999, 104, 1311–1320.
- Sanchez-Monge, R.; Lopez-Torrejon, G.; Pascual, C.Y.; Varela, J.; Martin-Esteban, M.; Salcedo, G. Vicilin and convicilin are potential major allergens from pea. Clin. Exp. Allergy 2004, 34, 1747–1753.
- Teufel, F.; Almagro Armenteros, J.J.; Johansen, A.R.; Gislason, M.H.; Pihl, S.I.; Tsirigos, K.D.; Winther, O.; Brunak, S.; von Heijne, G.; Nielsen, H. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat. Biotechnol. 2022, 40, 1023–1025.
- Wichers, H.J.; De Beijer, T.; Savelkoul, H.F.; Van Amerongen, A. The major peanut allergen Ara h 1 and its cleaved-off N-terminal peptide; possible implications for peanut allergen detection. J. Agric. Food Chem. 2004, 52, 4903–4907.
- Dubiela, P.; Kabasser, S.; Smargiasso, N.; Geiselhart, S.; Bublin, M.; Hafner, C.; Mazzucchelli, G.; Hoffmann-Sommergruber, K. Jug r 6 is the allergenic vicilin present in walnut responsible for IgE cross-reactivities to other tree nuts and seeds. Sci. Rep. 2018, 8, 11366.
- Goggin, D.E.; Mir, G.; Smith, W.B.; Stuckey, M.; Smith, P.M. Proteomic analysis of lupin seed proteins to identify conglutin Beta as an allergen, Lup an 1. J. Agric. Food Chem. 2008, 56, 6370–6377.
- Marcus, J.P.; Green, J.L.; Goulter, K.C.; Manners, J.M. A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 1999, 19, 699–710.
More
Information
Subjects:
Allergy
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
544
Revisions:
2 times
(View History)
Update Date:
06 Jun 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





