Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zunxi Huang | -- | 3354 | 2023-05-24 14:17:54 | | | |
| 2 | Dean Liu | -4 word(s) | 3350 | 2023-05-25 04:39:27 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Yang, Z.; Huang, Z.; Wu, Q.; Tang, X.; Huang, Z. Cold-Adapted Proteases. Encyclopedia. Available online: https://encyclopedia.pub/entry/44780 (accessed on 08 February 2026).
Yang Z, Huang Z, Wu Q, Tang X, Huang Z. Cold-Adapted Proteases. Encyclopedia. Available at: https://encyclopedia.pub/entry/44780. Accessed February 08, 2026.
Yang, Zhengfeng, Zhendi Huang, Qian Wu, Xianghua Tang, Zunxi Huang. "Cold-Adapted Proteases" Encyclopedia, https://encyclopedia.pub/entry/44780 (accessed February 08, 2026).
Yang, Z., Huang, Z., Wu, Q., Tang, X., & Huang, Z. (2023, May 24). Cold-Adapted Proteases. In Encyclopedia. https://encyclopedia.pub/entry/44780
Yang, Zhengfeng, et al. "Cold-Adapted Proteases." Encyclopedia. Web. 24 May, 2023.
Copy Citation
The modern biotechnology industry has a demand for macromolecules that can function in extreme environments. One example is cold-adapted proteases, possessing advantages such as maintaining high catalytic efficiency at low temperature and low energy input during production and inactivation. Meanwhile, cold-adapted proteases are characterised by sustainability, environmental protection, and energy conservation; therefore, they hold significant economic and ecological value regarding resource utilisation and the global biogeochemical cycle.
cold-adapted proteases
green energy conservation
stability
medical applications
1. Introduction
Proteases are a class of enzymes that can hydrolyse large proteins or peptides into amino acids or small peptides. The small peptides generated by hydrolysis often have unique physiological activity and are used in medicine, health care, and skin care products. Initial research on proteases can be traced back to the early 20th century; since then, with the rapid development of biotechnology, many researchers conducted comprehensive studies on proteases, making significant progress regarding strain screening, mechanism of action, structure-activity relationship, and the heterologous expression of protease. However, most previous research has focused on mesophilic proteases, while few studies have investigated cold-adapted proteases, which were mainly developed in the last 30 years. Cold-adapted proteases are commonly found in animals, plants, and microorganisms that are adapted to low-temperature environments. Most cold-adapted proteases have the optimal catalytic temperature of 10–40 °C and an optimal pH of 8–10. These alkalophilic proteases that can maintain high catalytic efficiency under low and medium temperatures and weakly alkaline conditions [1]. Approximately 80% of the earth’s biosphere and 90% of the marine environment experience temperatures below 5 °C [2]. The cold-adapted proteases used in industry mainly originate from extreme environments, including cold-adapted microorganisms in deep sea waters, polar regions, high mountains and plateaus, and cold deserts [1]. After long-term exposure to low temperature, cold adaptation microorganisms have evolved corresponding cold-adapted mechanisms, such as increased structural flexibility of enzymes, unique cell membrane lipid components, rapid cryoprotectant synthesis mechanisms, and high cold-shock protein expression through environmental adaptation and evolution, to ensure the fluidity of cell membranes and cell folding at low temperatures [3][4]. Compared with mesophilic proteases, cold-adapted proteases have a higher specific activity at low temperatures and require very low activation energies.
Researchers have discovered that psychrophilic microorganisms and cold-adapted enzymes have significant biotechnological potential. They provide many economic and ecological advantages compared with organisms and their enzymes that work at higher temperatures. For example, cold-adapted microorganisms and their cold-adapted proteins and enzymes have many biotechnological applications, in industries such as medicine, washing, textiles, food, waste management, and skin care [5]. Cold-adapted enzymes can be produced without heating, savings in energy and money. In industrial applications, the high activity of cold-adapted enzymes helps shorten the reaction times without a reduction in efficiency or auxiliary heating, thus also reducing the energy consumption rate [6]. For example, during leather production, cold-adapted proteases do not require additional heating and temperature adjustment, functioning at tap water temperatures compared with traditional mesophilic proteases. As a washing ingredient, cold-adapted proteases as a washing ingredient, this can be used to achieve decontamination at low temperatures, reducing the cost of environmental heating and avoiding possible colour damage resulting from high-temperature washing conditions [7]. In cold environments, the degradation ability of endogenous microorganisms decreases with decreasing temperature, which is not conducive to waste management in cold regions. However, cold-adapted proteases can compensate for this disadvantage. For example, treating organic wastes from fisheries and aquaculture can not only avoid serious health and environmental problems but can also recycle high-value biological molecules within waste sustainably [6].
Notably, proteases, as recognised therapeutic drugs with good tolerance, have been used in medicine for over a decade. Additionally, more than 100 years ago, proteases were used to treat chronic surface ulcers, tubulous lymphadenitis, and tubulous fibrosis [8][9][10]. Proteases are primarily used for treatment in four fields: oral medicine for gastrointestinal diseases, anti-infective drugs, thrombolytic drugs for thromboembolic diseases, and local drugs for wound debridement [11]. The majority, including thrombolytic drugs and coagulants, are used to treat blood diseases. Other approved protease therapies can be applied for digestion, muscle spasms, cosmetics, etc. such as using local proteases for selective tissue destruction to treat skin diseases (such as skin warts or actinic keratosis) [8].
Significant progress has recently been made in the research and application of industrial biocatalysis technology, although the potential of biocatalysis using extreme enzymes has not yet been fully realised [12]. Cold-adapted proteases are also more sensitive to thermal inactivation, low pH, and autolysis [13] while showing lower stability than mesophilic proteases in medium- and high-temperature environments. Improving the application and efficiency of cold-adapted proteases and their stability in medium- and high-temperature environments will be a primary direction for future research. Environmental problems such as climate change and global warming caused by greenhouse gas emissions call for humans to establish a low-carbon and sustainable green economy. Cold-adapted proteases are receiving increasing attention, and their related research, exploration, and application potential are also becoming more important.
2. Sources and Mining of Cold-Adapted Protease
It was previously believed that extremely cold areas were sterile and thus did not contain any life. Additionally, there was little interest in psychrophilic microorganisms that could grow at low temperatures. However, many microorganisms provide valuable molecular sources with high biotechnological potential [14]. As early as 1975, Morita defined psychrophilic bacteria based on their optimal growth temperatures. Their optimal growth temperature was determined to be 15 °C or lower, while the highest growth temperature was below 20 °C, and they could even grow when exposed to temperatures below 0 °C, differing from ordinary bacteria, whose optimum growth temperature is generally 20–4 °C [15][16]. During long-term exposure to low temperatures, cold-adapted microorganisms have evolved corresponding cold adaptation mechanisms, including increased structural flexibility of enzymes such as proteases, unique cell membrane lipid components, rapid cryoprotectant synthesis, and high cold-shock protein expression [7]. Cold temperatures are known to limit the movement of plants and animals. Thus, microorganisms play a dominant role in the cold environment, and their biological activities also maintain nutrient flow in cold environments while contributing to the global biogeochemical cycle [17].
Cold-adapted proteases primarily originate from cold-adapted microorganisms, which are generally found in extreme environments, such as water and mud in the deep sea, ice and frozen soil in polar regions, glaciers, mountains, and plateaus. The polar region covers an area of approximately 15%, with temperatures below 0 °C throughout the year. Glacial permafrost accounts for approximately 20% of the land area, with an average temperature below 4 °C. The oceans cover more than 70% of the earth, with an average temperature of 5 °C. These extreme environments are important habitats for cold-adapted microorganisms, which can be primarily divided into the following types: Gram-negative bacteria (Pseudoalteromonas, Moraxella, psychrophilic bacteria, Polar monospora, cold-resistant bacteria, etc.), gram-positive bacteria (Arthrobacter, Bacillus, Micrococcus, and Archaea, such as Methanogens, Rhodopseudomonas salina), yeast (Candida, Cryptococcus), and fungi (Penicillium, Cladosporium) [7].
The Himalayas are one of the most representative areas of low-temperature environments on Earth, with its bacterial diversity being recognised for its potential to produce low-temperature active enzymes. It has also been reported that there are temperature-sensitive and -tolerant bacterial isolates in the Qomolangma Glacier Site, indicating rich diversity even in the most extreme and harsh areas [17]. Furthermore, fungi on high-altitude glaciers are strong candidates for biotechnological applications, and according to a survey of the diversity of glacier fungi in Tirich Mir of the Dukush mountains in northern Pakistan, Penicillium is the most common, followed by Alternaria, which shows highly efficient low-temperature enzyme activity [18]. Park et al. [19] found 15,696 strains of bacteria that had been isolated from 633 seawater samples collected from the Chukchi Sea. Of these, 2526 strains (approximately 16%) were found to have protease activity after screening at a low temperature (15 °C). After 16S rRNA identification, these were found to have been primarily related to the following taxa: Alteromonas (31%), Staphylococcus (27%), Pseudoalteromonas (14%), Leeuwenhoecella (7%), Bacillus (5%), Thiobacillus (5%), cold-resistant bacilli (4%), Clostridium (2%), Acinetobacter (2%), Pseudomonas (1%), Halomonas (1%), and Dukes (1%). Additionally, Kim et al. [20] isolated 89 strains of bacteria from marine and land samples from the Svalbard Islands, Norway. Of these, 48 strains (approximately 54%), including Pseudoalteromonas (33 strains), Pseudomonas (10 strains), Arthrobacter (4 strains), and Flavobacterium (1 strain), were screened at low temperatures and observed to have protease activity.
Many psychrophilic enzymes, including psychrophilic proteases, have been discovered through different biomolecular and genetic engineering technologies. These have good application potential for different industrial applications and include microbial metagenomes from cryogenic environments, psychrophilic enzymes obtained from various cold environments, including proteases [20] and cellulases [21][22], xylanases [23], amylases [24], lipases [25], chitinases [26], pectinases [27], esterases [28][29], and laccases [30]. Three proteases and other enzymes of important commercial value from Antarctic soil were previously obtained using the metagenomic method [31]. Fan et al. successfully obtained a new cold-adapted N-acyl homoserine lactoesterase from the macro microbe genome of musty tofu, which could significantly reduce the production of virulence factors and biofilms of Pseudomonas aeruginosa PAO1 and also has the potential to become a candidate drug for treating P. aeruginosa infections [32]. Furthermore, Tchigvintsev et al. successfully screened several carboxylesterases from the marine metagenome, including some with excellent cold adaptation and salt tolerance [33].
Metagenomics, as a mining method for discovering new enzyme genes in microorganisms and their communities, bypasses the technical challenge of cultivating polar microorganisms and has shown excellent application potential in many related reports. Screening environmental microbial strains is also an effective method for mining enzyme genes. This method can obtain enzyme genes with special functions through the enrichment and screening of culturable microorganisms in the corresponding environment and is characterised by being fast, simple, and suitable for large-scale screening. Arnórsdottir et al. previously reported that a cold-adapted protease Vpr had been obtained from a marine psychrophilic vibrio, and high expression was achieved using the T7 system of Escherichia coli [34][35]. Additionally, Lario et al. studied a protease from cold-adapted yeast from Antarctica before purifying and sequencing it [36]. A protease produced by cold nutritious Bacillus fetida was isolated from Himalayan glacial soils. Through qualitative and quantitative screening, the cold-active protease Apr-BO1 was purified, which can effectively remove stains during low-temperature washing and is considered a good detergent additive [37]. Daskaya-Dikmen et al. screened yeast strains of different genera from a cold environment in northern Turkey and screened for cold-adapted pectinases, proteases, amylases, etc. Among them, the production of cold-active amylase by Cystosporium capitatum and pectinase by Rhodosporomyces was first reported [38]. The comparative study of functional genomics is another efficient and fast method of gene mining. With the progress of high-throughput sequencing technology, the number of publicly available bacterial genomes has increased significantly. Through the comparative analysis of genome sequences, researchers found genes encoding biological molecules and products of interest. Perfumo et al. used the comparative genomics of Psychrobacter genes to specifically identify and clone a gene that encoded a cold-active protease in isolate 94–6 PB and subsequently characterised the physical and chemical properties of the expression enzyme through in vitro analysis [39].
3. Classification of Cold-Adapted Proteases
The conventional classification method for proteases can also be applied to classify cold-adapted proteases. Proteases can be divided into acidic, alkaline, and neutral categories according to the degree of acid–base preference. According to the method of peptide bond hydrolysis, these can be divided into endopeptidases (hydrolysis of internal peptide bonds) and exopeptidases (hydrolysis from the end). Endopeptidases are widely used in industries, depending on the chemical properties of functional groups in catalytic or active sites and can be generally divided into serine proteases, metalloproteinases, cysteine proteases, and aspartate proteases. They can be further classified according to the degree of inhibition of enzyme inhibitors [21] into serine protease inhibitors (benzene sulfonyl fluoride PMSF), metalloproteinase inhibitors (1,10-phenanthroline and EDTA), cysteine protease inhibitors (N-ethylmaleimide), and aspartic protease inhibitors (pepsin inhibitor A). PMSF can covalently bind with serine residues to cause conformational changes in the enzyme molecule; the more serine residues in the active part of the protease, the stronger the inhibition of PMSF on the enzyme. Serine proteases are widely distributed across various taxa, indicating that serine proteases are crucial for the survival of microorganisms [40].
If the protease activity decreases by more than 20% after treatment with inhibitors, which constitutes a significant negative impact, the protease can be classified according to the degree of inhibition. Inhibitor analysis of proteases at low temperatures can determine the type of cold-adapted protease. PMSF reduced the activity of ArcP02, ArcP08, and ArcP11 protease crude enzyme solutions by 24.8%, 39.8%, and 31.4%, respectively, whereas those of ArcP02 and ArcP08 decreased by 27.3% and 21.2%, respectively, at the same concentration of 1,10-phenanthroline. Therefore, it can be considered that the proteases produced by ArcP02, ArcP08, and ArcP11 are serine proteases, whilst ArcP02 and ArcP08 may produce additional metalloproteinases [19]. Table 1 shows that a large proportion of the reported cold-adapted proteases belong to serine proteases [12][40][41][42][43][44][45][46][47][48][49][50], and a part of them belongs to metalloproteinases [51][52]. According to the optimal pH, most cold-adapted proteases belong to neutral proteases [40][50][51][53][54][55] and alkaline proteases [41][42][43][44][45][46][56][57][58][59]. In contrast, very few belong to acid proteases [12][60] because the activity of acid cold-adapted proteases contains aspartic acid residues; therefore, they are also aspartic proteases. In general, cold-adapted proteases are known to be active against various natural proteins and different types of natural and synthetic substrates, showing a wide range of substrate specificity. The cold-adapted serine proteases produced by Chryseobacterium sp. have the highest hydrolytic activity against casein, followed by gelatin and bovine serum albumin, and the lowest on egg albumin [40]. The cold-adapted serine peptidase produced by Lysobacter sp. has the highest activity against azocasein, followed by gelatin and feather powder, and the lowest activity against casein, bovine serum albumin, and azo keratin [52]. The broad substrate specificity of cold-adapted proteases is of great value in industrial applications, especially in bioremediation processes performed at low temperatures.
Table 1. Enzymatic properties of cold-adapted protease.
| Source/Protease | Temp Optima (°C) |
pH Optima |
Molecular Weight (kDa) |
Inhibitors | Activators | Refs. |
|---|---|---|---|---|---|---|
| Alteromonas haloplanktis | 20 | 8.0–9.0 | 74–76 | Leupeptin, PCMB | - | [57] |
| Aspergillus ustus | 45 | 9.0 | 32 | PMSF, DFP, Cu2+ | - | [45] |
| Bacillus subtilis PAMC 26541 | 40 | 7.0–7.5 | 107 | PMSF | K+, Na+ | [50] |
| Bacillus sp. S1DI 10 | 10 | 8 | 40 | EDTA, PMSF, Leucopeptin | Fe2+, Mn2+, Co2+, Twain 80 | [41] |
| Bacillus subtilis WLCP1 | 15 | 10 | 38 | PMSF | Ca2+, Cu2+ | [49] |
| Bacillus cereus | 20 | 9.0 | - | EDTA, PMSF, Ca2+, Cu2+, K+ | Co2+, Fe2+ | [46] |
| Bacillus cereus | 42 | 7.0–8.5 | 34.2 | Mg2+, Mn2+ | - | [58] |
| Curtobacterium luteum | 20 | 7 | 115 | EDTA, EGTA | Zn2+, Cr2+ | [51] |
| Colwellia sp. NJ341 | 35 | 8.0–9.0 | 60 | PMSF, Fe2+ | - | [43] |
| Colwellia psychrerythraea | 19 | 6–8.5 | 71 | EDTA, Zn2+, Mn2+ | Na+, Mg2+ | [55] |
| Candida humicola | 37 | 1–1.2 | 36 | - | - | [60] |
| Chryseobacterium sp. IMDY | 10 | 7.0–8.0 | 27 | PMSF, Hg2+, Zn2+ | Na+, Ca2+ | [40] |
| Escherichia freundii | 25 | 10.0 | 55 | Iodoacetamide, SDS |
- | [61] |
| Flavobacterium psychrophilum |
24 | 6.0–7.0 | 62 | Benzalkonium chloride, Na+, Phenanthroline, | - | [53] |
| Pseudomonas lundensis |
40 | 10.4 | 46 | EDTA, PMSF Cu2+, Zn2+, Hg2+, EDTA, SDS |
Mg2+, Ca2+ | [56] |
| Halobillus sp. SCSIO 20089 | 30 | 8 | 35 | EDTA | Ca2+, Mg2+, Mn2+ | [55] |
| Leucosporidium antarcticum |
25 | 6.7–7.1 | 34.4 | - | - | [54] |
| Lysobacter sp. | 40 | 9.0 | 35 | PMSF, EDTA, Zn2+ | Ca2+, Mg2+, Ba2+, Na+, NH4+, isopropyl alcohol | [52] |
| Penicillium chrysogenum FS010 | 35 | 9 | 41 | PMSF, DFP, Cu2+, Co2+ | Mg2+, Ca2+ | [42] |
| Psychrobacter sp. 94–6 PB | 30 | 9 | 80 | - | - | [39] |
| Planococcus sp. M7 | 35 | 10 | 43 | PMSF, TNBS, EDAC, EDTA, Cu2+, Ni2+ | Fe3+, Ca2+ | [44] |
| Pseudomonas DY | 40 | 10 | 25 | PMSF, DFP, AEBSF | Ca2+, Mg2+ | [43] |
| Pseudoalteromonas arctica PAMC 21717 | 30 | 9 | 37 | LAS, SDS | Ca2+ | [59] |
| Pedobacter cryoconitis | 40 | 8.0 | 27 | - | - | [62] |
| Pseudoaltermonas. sp. | 30 | 8.0 | 47 | PMSF, Chymostatin, Trypsin |
- | [63] |
| Pseudomonas strain DY-A | 40 | 10.0 | - | EDTA, EGTA, SDS | - | [47] |
| Stenotrophomonas IIIM-ST045 | 15 | 10 | 55 | Zn2+, Cu2+, Co2+ | Mg2+, Mn2+, Ca2+ | [64] |
| Serratia marcescens | 40 | 6.5–8.0 | 58 | Phenanthroline, PSFM, EGTA, DTT | - | [48] |
| Sphingomonas paucimobilis |
20–30 | 6.5–7.0 | - | DFP, PMSF, AEBSF | - | [65] |
| Trichoderma Atroviride |
25 | 6.2 | 24 | SDS, Urea | - | [12] |
4. Enzymatic Characteristics of Cold-Adapted Proteases
For normal growth adaptation to life under low-temperature conditions, cold-adapted microorganisms have evolved a set of gene expression sequences and special molecular mechanisms specific to low-temperature environments, in which the high expression of low-temperature active enzymes plays a key role during the process of cold adaptation. In recent decades, research on cold-adapted proteases has initially focused on exploring enzymatic characteristics, including the optimal temperature, pH, and Effects of metal ions (e.g., Hg2+, Cd2+, Co2+, Mn2+, Ca2+, K+, Na+, Fe2+, etc.) and compounds (such as SDS, PMSF, EDTA, LAS, etc.) on enzyme activity. Their optimal catalytic temperature is 10–60 °C, whilst their optimal pH is usually in the neutral to alkaline pH range of 7–10 [1]. To date, thermostable alkaline proteases primarily originate from Bacillus subtilis and Bacillus licheniformis, whilst proteases from Geomyces panorum, and yeast Candida humicola are rare acid thermostable proteases with an optimal pH of 3.0 [1][56][60]. Presently, the reported molecular weight of cold-adapted proteases is small, typically in the range of 25–60 kDa; only a few cold-adapted proteases have large molecular weights [39][50][51][57]. Organic reagents such as EDTA, PMSF, and DFP often inhibit the activity of cold-adapted protease [41][42][43], as do metal ions Hg2+, Cu2+ and Co2+ [40][66]. Metal ions, such as Mg2+, Mn2+, and Ca2+ can improve the activity of cold-adapted proteases to varying degrees [46][58].
5. Structural Adaptations of Cold-Adapted Protease
The structure of cold-adapted proteases can be analysed from their molecular (amino acid composition and sequence, terminal composition, disulphide bond position, etc.) and three-dimensional structure. Nuclear magnetic resonance spectroscopy, site-directed mutagenesis, X-ray crystal diffraction, and freeze electron microscopy are effective methods that can be used to understand the crystal structure. Additionally, the high structural homology of enzymes at different temperatures provides an opportunity to study the structural characteristics of their adaptation to different temperatures. Through comparative analysis of the structural characteristics of cold-adapted, mesophilic, and thermophilic proteases (Figure 1), as well as previous studies, researchers can understand how proteases become adapted to low temperatures. These groups comprise protein surfaces that determine the important interactions between proteins and water, which is expected to significantly impact protein function in adapting to high and low temperatures. In many thermophilic proteins, a larger proportion of non-polar surfaces is believed to help increase stability [67][68]. The proportion of serine residues in cold-adapted enzyme molecules of cold-adapted microorganisms is high, while the proportion of proline and acid residues is low. Owing to the high number of polar amino acid residues, these types of enzymes have a more hydrophilic interaction with solvents [49][69]. Therefore, the active sites of these enzymes are usually more accessible to compensate for substrate diffusion at lower temperatures. The difference between cold-adapted Vibrio proteases and more stable proteases is their strong anionic properties due to the numerous uncompensated negatively-charged residues on their surface. In some studies, the difference in surface charge distribution or the increase in the non-polar surface area has been considered to enable adaptation to low temperatures [70][71][72]. On the other hand, the weakness of non-covalent interactions (such as salt bridges, hydrophobic interactions, aromatic interactions, main chains, side chains, and side chain hydrogen bonds) in some special positions can increase the core volume of enzyme molecules and make the whole molecule looser [46], improving the conformational flexibility of proteases [37]. However, the relationship between the number of non-covalent bonds and the temperature adaptation of enzymes cannot be determined at present. For example, the number of salt bridges in cold-adapted protease 1SH7, thermophilic protease 1IC6, and thermophilic protease 1THM was compared [73]. The number of salt bridges in 1SH7 and 1IC6 was the same and was only two less than that in thermophilic protease 1THM. Although there have been many reported instances of salt bridges and hydrogen bonds introduced in the modification of thermal stability enhancement of many enzymes, an important aspect of their contribution to protein stability lies in their location and distribution [73]. However, this increase in flexibility is often accompanied by decreased stability. Cold-adapted enzymes generally show low thermal stability, short half-life, and high sensitivity to low pH [74]. In contrast, thermostable enzymes have a more rigid and compact conformation, which is more likely to protect them from instability at higher temperatures [75].
From a kinetic point of view, a very common feature of all low-temperature active enzymes is the reduction of activation enthalpy, accompanied by negative entropy, which reduces the exponential decline of the chemical reaction rate resulting from the reduction in temperature. It has been reported that the surface flexibility of low-temperature active enzymes is the source of cold adaptability [76]. For example, Isaksen discussed the role of protein surface mobility in determining the enthalpy centre equilibrium and subsequently proved that the surface of the enzyme is rigid through a single remote mutation of the key residue of protein water surface interaction. Therefore, cold-denatured trypsin can be transformed into a variant with medium-temperature characteristics without changing the amino acid sequence [77]. It has been suggested that the rigidity of the surface of low-temperature active enzymes may produce a higher activation enthalpy of the catalytic reaction with fewer negative entropy components.
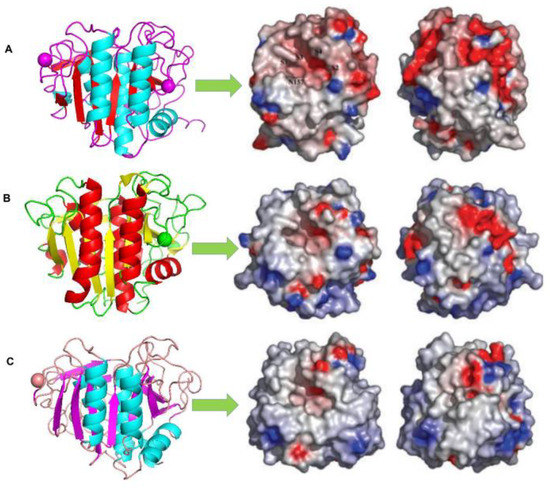
Figure 1. Comparison of the electrostatic surface potentials and crystal structures of (A) 1SH7, (B) 1IC6, and (C) 1THM [73]. On the right-hand side, the molecules have been rotated 180 about the y-axis. The approximate locations of substrate binding pockets, S1–S4 (nomenclature according to) and the oxyanion hole residue, N157, are labelled on the surface of the Vibrio proteinase (A) [78]. The positive potential is in blue and the negative potential is in red. The electrostatic surface potential was calculated using Delphi, and the graphical presentations were produced using Pymol [63].
References
- Furhan, J. Adaptation, production, and biotechnological potential of cold-adapted proteases from psychrophiles and psychrotrophs: Recent overview. J. Genet. Eng. Biotechnol. 2020, 18, 36.
- Kumar, A.; Mukhia, S.; Kumar, R. Industrial applications of cold-adapted enzymes: Challenges, innovations and future perspective. 3 Biotech 2021, 11, 426.
- Collins, T.; Margesin, R. Psychrophilic lifestyles: Mechanisms of adaptation and biotechnological tools. Appl. Microbiol. Biotechnol. 2019, 103, 2857–2871.
- Shen, l.-J.; Zhang, S.-T.; Chen, G. Regulated strategies of cold-adapted microorganisms in response to cold: A review. Environ. Sci. Pollut. Res. 2021, 28, 68006–68024.
- Gounot, A.M. Bacterial life at low temperature: Physiological aspects and biotechnological implications. J. Appl. Bacteriol. 1991, 71, 386–397.
- Khiari, Z. Sustainable Upcycling of Fisheries and Aquaculture Wastes Using Fish-Derived Cold-Adapted Proteases. Front. Nutr. 2022, 9, 875697.
- Nielsen, M.H.; Jepsen, S.J.; Outtrup, H. Enzymes for low temperature washing. J. Am. Oil Chem. Soc. 1981, 58, 644–649.
- Craik, C.S.; Page, M.J.; Madison, E.L. Proteases as therapeutics. Biochem. J. 2011, 435, 1–16.
- Morani, A.D. Trypsin therapy in the management of chronic surface ulcers. Plast. Reconstr. Surg. 1953, 11, 372–379.
- Rapoport, C. The use of trypsin in the therapy of tuberculous lymphadenitis and tuberculous fistulae. Dis. Chest. 1958, 34, 154–161.
- Gudmundsdottir, A.; Palsdottir, H.M. Atlantic cod trypsins: From basic research to practical applications. Mar. Biotechnol. 2005, 7, 77–88.
- Sarmiento, F.; Peralta, R.; Blamey, J.M. Cold and Hot Extremozymes: Industrial Relevance and Current Trends. Front. Bioeng. Biotechnol. 2015, 3, 148.
- Fornbacke, M.; Clarsund, M. Cold-adapted proteases as an emerging class of therapeutics. Infect. Dis. Ther. 2013, 2, 15–26.
- Perfumo, A.; Banat, I.M.; Marchant, R. Going Green and Cold: Biosurfactants from Low-Temperature Environments to Biotechnology Applications. Trends Biotechnol. 2018, 36, 277–289.
- Morita, R.Y. Psychrophilic bacteria. Bacteriol. Rev. 1975, 39, 144–167.
- Kralova, S. Role of fatty acids in cold adaptation of Antarctic psychrophilic Flavobacterium spp. Syst. Appl. Microbiol. 2017, 40, 329–333.
- Dhakar, K.; Pandey, A. Microbial Ecology from the Himalayan Cryosphere Perspective. Microorganisms 2020, 8, 257.
- Rafiq, M.; Nadeem, S.; Hassan, N.; Hayat, M.; Sajjad, W.; Zada, S.; Sajjad, W.; Hasan, F. Fungal recovery and characterization from Hindu Kush mountain range, Tirich Mir glacier, and their potential for biotechnological applications. J. Basic. Microbiol. 2020, 60, 444–457.
- Park, H.J.; Lee, Y.M.; Kim, S.; Wi, A.R.; Han, S.J.; Kim, H.W.; Kim, I.C.; Yim, J.H.; Kim, D. Identification of proteolytic bacteria from the Arctic Chukchi Sea expedition cruise and characterization of cold-active proteases. J. Microbiol. 2014, 52, 825–833.
- Kim, E.H.; Cho, K.H.; Lee, Y.M.; Yim, J.H.; Lee, H.K.; Cho, J.C.; Hong, S.G. Diversity of cold-active protease-producing bacteria from arctic terrestrial and marine environments revealed by enrichment culture. J. Microbiol. 2010, 48, 426–432.
- Bhat, A.; Riyaz-Ul-Hassan, S.; Ahmad, N.; Srivastava, N.; Johri, S. Isolation of cold-active, acidic endocellulase from Ladakh soil by functional metagenomics. Extremophiles 2013, 17, 229–239.
- Oh, H.N.; Park, D.; Seong, H.J.; Kim, D.; Sul, W.J. Antarctic tundra soil metagenome as useful natural resources of cold-active lignocelluolytic enzymes. J. Microbiol. 2019, 57, 865–873.
- Paixao, D.; Tomazetto, G.; Sodre, V.R.; Goncalves, T.A.; Uchima, C.A.; Buchli, F.; Alvarez, T.M.; Persinoti, G.F.; Da, S.M.; Bragatto, J.; et al. Microbial enrichment and meta-omics analysis identify Cazymes from mangrove sediments with unique properties. Enzyme Microb. Technol. 2021, 148, 109820.
- Kim, Y.D.; Kim, J.H.; Lee, Y.M.; Lee, J.S.; Shin, D.H.; Ku, B.H.; Son, K.H.; Park, H.Y. Identification and Characterization of a Novel, Cold-Adapted D-Xylobiose- and D-Xylose-Releasing Endo-β-1,4-Xylanase from an Antarctic Soil Bacterium, Duganella sp. PAMC 27433. Biomolecules 2021, 11, 680.
- Kumar, R.; Acharya, V.; Mukhia, S.; Singh, D.; Kumar, S. Complete genome sequence of Pseudomonas frederiksbergensis ERDD5:01 revealed genetic bases for survivability at high altitude ecosystem and bioprospection potential. Genomics 2019, 111, 492–499.
- Liu, K.; Ding, H.-T.; Yu, Y.; Chen, B. A Cold-Adapted Chitinase-Producing Bacterium from Antarctica and Its Potential in Biocontrol of Plant Pathogenic Fungi. Mar. Drugs. 2019, 17, 695.
- Nakagawa, T.; Nagaoka, T.; Taniguchi, S.; Miyaji, T.; Tomizuka, N. Isolation and characterization of psychrophilic yeasts producing cold-adapted pectinolytic enzymes. Lett. Appl. Microbiol. 2004, 38, 383–387.
- See-Too, W.S.; Convey, P.; Pearce, D.A.; Chan, K.G. Characterization of a novel N-acylhomoserine lactonase, AidP, from Antarctic Planococcus sp. Microb. Cell Fact. 2018, 17, 179.
- Kryukova, M.V.; Petrovskaya, L.E.; Kryukova, E.A.; Lomakina, G.Y.; Yakimov, S.A.; Maksimov, E.G.; Boyko, K.M.; Popov, V.O.; Dolgikh, D.A.; Kirpichnikov, M.P. Thermal Inactivation of a Cold-Active Esterase PMGL3 Isolated from the Permafrost Metagenomic Library. Biomolecules 2019, 9, 880.
- Yang, Q.; Zhang, M.; Zhang, M.; Wang, C.; Liu, Y.; Fan, X.; Li, H. Characterization of a Novel, Cold-Adapted, and Thermostable Laccase-Like Enzyme with High Tolerance for Organic Solvents and Salt and Potent Dye Decolorization Ability, Derived from a Marine Metagenomic Library. Front. Microbiol. 2018, 9, 2998.
- Berlemont, R.; Pipers, D.; Delsaute, M.; Angiono, F.; Feller, G.; Galleni, M.; Power, P. Exploring the Antarctic soil metagenome as a source of novel cold-adapted enzymes and genetic mobile elements. Rev. Argent. Microbiol. 2011, 43, 94–103.
- Fan, X.J.; Liang, M.J.; Wang, L.; Chen, R.; He, L.; Liu, X.L. Aii810, a Novel Cold-Adapted N-Acylhomoserine Lactonase Discovered in a Metagenome, Can Strongly Attenuate Pseudomonas aeruginosa Virulence Factors and Biofilm Formation. Front. Microbiol. 2017, 8, 1950.
- Tchigvintsev, A.; Tran, H.; Popovic, A.; Kovacic, F.; Brown, G.; Flick, R.; Hajighasemi, M.; Egorova, O.; Somody, J.C.; Tchigvintsev, D.; et al. The environment shapes microbial enzymes: Five cold-active and salt-resistant carboxylesterases from marine metagenomes. Appl. Microbiol. Biol. 2015, 99, 2165–2178.
- Arnórsdottir, J.; Smáradóttir, R.B.; Magnússon, O.T.; Thorbjarnardóttir, S.H.; Eggertsson, G.; Kristjánsson, M.M. Characterization of a cloned subtilisin-like serine proteinase from a psychrotrophic Vibrio species. Eur. J. Biochem. 2002, 269, 5536–5546.
- Oskarsson, K.R.; Kristjansson, M.M. Improved expression, purification and characterization of VPR, a cold active subtilisin-like serine proteinase and the effects of calcium on expression and stability. Biochim. Biophys. Acta Proteins Proteom. 2019, 1867, 152–162.
- Lario, L.D.; Pillaca-Pullo, O.S.; Duraes, S.L.; Converti, A.; Casati, P.; Spampinato, C.; Pessoa, A. Optimization of protease production and sequence analysis of the purified enzyme from the cold adapted yeast Rhodotorula mucilaginosa CBMAI 1528. Biotechnol. Rep. 2020, 28, e546.
- Farooq, S.; Nazir, R.; Ganai, S.A.; Ganai, B.A. Isolation and characterization of a new cold-active protease from psychrotrophic bacteria of Western Himalayan glacial soil. Sci. Rep. 2021, 11, 12768.
- Daskaya-Dikmen, C.; Karbancioglu-Guler, F.; Ozcelik, B. Cold active pectinase, amylase and protease production by yeast isolates obtained from environmental samples. Extremophiles 2018, 22, 599–606.
- Perfumo, A.; Freiherr, V.S.G.; Nordmann, E.L.; Budisa, N.; Wagner, D. Discovery and Characterization of a New Cold-Active Protease From an Extremophilic Bacterium via Comparative Genome Analysis and in vitro Expression. Front. Microbiol. 2020, 11, 881.
- Mageswari, A.; Subramanian, P.; Chandrasekaran, S.; Karthikeyan, S.; Gothandam, K.M. Systematic functional analysis and application of a cold-active serine protease from a novel Chryseobacterium sp. Food Biochem. 2017, 217, 18–27.
- Singh, D.; Thakur, S.; Thayil, S.M.; Kesavan, A.K. Characterization of a cold-active, detergent-stable metallopeptidase purified from Bacillus sp. S1DI 10 using Response Surface Methodology. PLoS ONE. 2019, 14, e216990.
- Zhu, H.Y.; Tian, Y.; Hou, Y.H.; Wang, T.H. Purification and characterization of the cold-active alkaline protease from marine cold-adaptive Penicillium chrysogenum FS010. Mol. Biol. Rep. 2009, 36, 2169–2174.
- Wang, Q.F.; Miao, J.L.; Hou, Y.H.; Ding, Y.; Wang, G.D.; Li, G.Y. Purification and characterization of an extracellular cold-active serine protease from the psychrophilic bacterium Colwellia sp. NJ341. Biotechnol. Lett. 2005, 27, 1195–1198.
- Chen, K.; Mo, Q.; Liu, H.; Yuan, F.; Chai, H.; Lu, F.; Zhang, H. Identification and characterization of a novel cold-tolerant extracellular protease from Planococcus sp. CGMCC 8088. Extrenophiles 2018, 22, 473–484.
- Damare, C.; Raghukuma, C.; Muraleedharan, U.D.; Raghukumar, S. Deep-sea fungi as a source of alkaline and cold-tolerant proteases. Enzyme Microb. Technol. 2006, 39, 172–181.
- Joshi, G.K.; Kumar, S.; Sharma, V. Production of moderately halotolerant, SDS stable alkaline protease from Bacillus cereus MTCC 6840 isolated from lake Nainital, Uttaranchal state, India. Braz. J. Microbiol. 2007, 38, 773–779.
- Zeng, R.; Zhang, R.; Zhao, J.; Lin, N. Cold-active serine alkaline protease from the psychrophilic bacterium Pseudomonas strain DY-A: Enzyme purification and characterization. Extremophiles 2003, 7, 335–337.
- Morita, Y.; Kondoh, K.; Hasan, Q.; Sakaguchi, T.; Murakami, Y.; Yokoyama, K.; Tamiya, E. Purification and characterization of a cold-active protease from psychrotrophic Serratia marcescens AP3801. J. Am. Oil Chem. Soc. 1997, 74, 1377–1383.
- Raymond-Bouchard, I.; Goordial, J.; Zolotarov, Y.; Ronholm, J.; Stromvik, M.; Bakermans, C.; Whyte, L.G. Conserved genomic and amino acid traits of cold adaptation in subzero-growing Arctic permafrost bacteria. FEMS Microbiol. Ecol. 2018, 94.
- Kim, H.D.; Kim, S.M.; Choi, J.I. Purification, Characterization, and Cloning of a Cold-Adapted Protease from Antarctic Janthinobacterium lividum. J. Microbiol. Biotechn. 2018, 28, 448–453.
- Kuddus, M.; Ramteke, P.W. A cold-active extracellular metalloprotease from Curtobacterium luteum (MTCC 7529): Enzyme production and characterization. J. Gen. Appl. Microbiol. 2008, 54, 385–392.
- Pereira, J.Q.; Ambrosini, A.; Passaglia, L.M.P.; Brandelli, A. A new coldadapted serine peptidase from Antarctic Lysobacter sp. A03: Insights about enzyme activity at low temperatures. Int. J. Biol. Macromol. 2017, 103, 854–862.
- Secades, P.; Alvarez, B.; Guijarro, J.A. Purification and properties of a new psychrophilic metalloprotease (Fpp2) in the fish pathogen Flavobacterium psychrophilum. FEMS Microbiol. Lett. 2003, 226, 273–279.
- Turkiewicz, M.; Pazgier, M.; Kalinowska, H.; Bielecki, S. A cold-adapted extracellular serine proteinase of the yeast Leucosporidium antarcticum. Extremophiles 2003, 7, 435–442.
- Singh, P.; Singh, S.M.; Dhakephalkar, P. Diversity, cold active enzymes and adaptation strategies of bacteria inhabiting glacier cryoconite holes of High Arctic. Extremophiles 2014, 18, 229–242.
- Yang, C.; Wang, F.; Hao, J.; Zhang, K.; Yuan, N.; Sun, M. Identification of a proteolytic bacterium, HW08, and characterization of its extracellular coldactive alkaline metalloprotease Ps5. Biosci. Biotechnol. Biochem. 2010, 74, 1220–1225.
- Suzuki, S.; Odagami, T. Low-temperature-active thiol protease from marine bacterium Alteromonas haloplanktis. J. Biotechnol. 1997, 5, 230–233.
- Shi, J.S.; Wu, Q.F.; Xu, Z.H.; Tao, W.Y. Identification of psychrotrophs SYP-A2-3 producing cold-adapted protease from the No. 1 Glacier of China and study on its fermentation conditions. Wei Sheng Wu Xue Bao 2005, 45, 258–263.
- Park, H.J.; Lee, C.W.; Kim, D.; Do, H.; Han, S.J.; Kim, J.E.; Koo, B.H.; Lee, J.H.; Yim, J.H. Crystal structure of a cold-active protease (Pro21717) from the psychrophilic bacterium, Pseudoalteromonas arctica PAMC 21717, at 1.4 A resolution: Structural adaptations to cold and functional analysis of a laundry detergent enzyme. PLoS ONE 2018, 13, e191740.
- Ray, M.K.; Devi, K.U.; Kumar, G.S.; Shivaji, S. Extracellular protease from the antarctic yeast Candida humicola. Appl. Environ. Microbiol. 1992, 58, 1918–1923.
- Nakajima, M.; Mizusawa, K.; Yoshida, F. Purification and properties of an extracellular proteinase of psychrophilic Escherichia freundii. Eur. J. Biochem. 1974, 44, 87–96.
- Margesin, R.; Dieplinger, H.; Hofmann, J.; Sarg, B.; Lindner, H. A cold-active extracellular metalloprotease from Pedobacter cryoconitis-production and properties. Res. Microbiol. 2005, 156, 499–505.
- Honig, B.; Nicholls, A. Classical electrostatics in biology and chemistry. Science 1995, 268, 1144–1149.
- Saba, I.; Qazi, P.H.; Rather, S.A.; Dar, R.A.; Qadri, Q.A.; Ahmad, N.; Johri, S.; Taneja, S.C.; Shawl, S. Purification and characterization of a cold active alkaline protease from Stenotrophomonas sp. isolated from Kashmir, India. World J. Microb. Biot. 2012, 28, 1071–1079.
- Turkiewicz, M.; Gromek, E.; Kalinowska, H.; Zielinska, M. Biosynthesis and properties of an extracellular metalloprotease from the Antarctic marine bacterium Sphingomonas paucimobilis. J. Biotechnol. 1999, 70, 53–60.
- Yang, J.; Li, J.; Mai, Z.; Tian, X.; Zhang, S. Purification, characterization, and gene cloning of a cold-adapted thermolysin-like protease from Halobacillus sp. SCSIO 20089. J. Biosci. Bioeng. 2013, 115, 628–632.
- Szilagyi, A.; Zavodszky, P. Structural differences between mesophilic, moderately thermophilic and extremely thermophilic protein subunits: Results of a comprehensive survey. Structure 2000, 8, 493–504.
- Haney, P.J.; Badger, J.H.; Buldak, G.L.; Reich, C.I.; Woese, C.R.; Olsen, G.J. Thermal adaptation analyzed by comparison of protein sequences from mesophilic and extremely thermophilic Methanococcus species. Proc. Natl. Acad. Sci. USA 1999, 96, 3578–3583.
- Bialkowska, A.M.; Morawski, K.; Florczak, T. Extremophilic proteases as novel and efficient tools in short peptide synthesis. J. Ind. Microbiol. Biol. 2017, 44, 1325–1342.
- Aghajari, N.; Feller, G.; Gerday, C.; Haser, R. Structures of the psychrophilic Alteromonas haloplanctis alpha-amylase give insights into cold adaptation at a molecular level. Structure 1998, 6, 1503–1516.
- Kim, S.Y.; Hwang, K.Y.; Kim, S.H.; Sung, H.C.; Han, Y.S.; Cho, Y. Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum. J. Biol. Chem. 1999, 274, 11761–11767.
- Russell, R.J.; Gerike, U.; Danson, M.J.; Hough, D.W.; Taylor, G.L. Structural adaptations of the cold-active citrate synthase from an Antarctic bacterium. Structure 1998, 6, 351–361.
- Arnorsdottir, J.; Kristjansson, M.M.; Ficner, R. Crystal structure of a subtilisin-like serine proteinase from a psychrotrophic Vibrio species reveals structural aspects of cold adaptation. FEBS J. 2005, 272, 832–845.
- Siddiqui, K.S.; Cavicchioli, R. Cold-adapted enzymes. Annu. Rev. Biochem. 2006, 75, 403–433.
- Miyazaki, K.; Wintrode, P.L.; Grayling, R.A.; Rubingh, D.N.; Arnold, F.H. Directed evolution study of temperature adaptation in a psychrophilic enzyme. J. Mol. Biol. 2000, 297, 1015–1026.
- Isaksen, G.V.; Aqvist, J.; Brandsdal, B.O. Protein surface softness is the origin of enzyme cold-adaptation of trypsin. PLoS Comput. Biol. 2014, 10, e1003813.
- Isaksen, G.V.; Aqvist, J.; Brandsdal, B.O. Enzyme surface rigidity tunes the temperature dependence of catalytic rates. Proc. Natl. Acad. Sci. USA 2016, 113, 7822–7827.
- Matthews, B.W. Solvent content of protein crystals. J. Mol. Biol. 1968, 33, 491–497.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
773
Revisions:
2 times
(View History)
Update Date:
25 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




