1. Introduction
RNA technology has existed for decades
[1]. However, its validation came with the fast spread of COVID-19
[2], which usually takes years to achieve
[3]. While several RNA products had been approved, a vaccine that delivers an antigen has resulted in a new focus on future therapies, from the creation of vaccines against infections to preventing or treating autoimmune disorders, supplying proteins by creating an innate manufacturing facility, assisting in gene editing, and many more novel applications
[4]. Technological and regulatory advancements include more efficient and lower-cost GMP manufacturing, cell-free DNA sourcing, improved purity profiles, and validated analytical methods from the United States Pharmacopoeia. The realization that we can now treat thousands of untreatable diseases represents a paradigm shift that will bring many ways to utilize nucleoside medicines.
2. Approved Therapies
The approved nucleic acid (DNA/RNA) therapeutics treat diseases by targeting their genetic blueprints in vivo, unlike targeting proteins, which is a conventional transient approach. The long-term curative effects of nucleic acid therapies are driven by gene inhibition, addition, replacement, or editing. Unlike other treatment techniques, nucleic acid therapies’ efficacy and applicability in the last few decades have depended on delivery technologies that have enhanced stability, facilitated internalization, and boosted target affinity. Nucleic acid therapeutics include antisense oligonucleotides, ligand-modified small interfering RNA conjugates, lipid nanoparticles, and adeno-associated virus vectors.
Non-coding RNA therapies include short oligonucleotides that bind to complementary sequences in endogenous RNA transcripts and change their processing to replace faulty proteins or existing vaccine antigens. ASO stands for antisense oligonucleotide, a steric block that can prevent polyadenylation, impede or promote translation, or change splicing by physically inhibiting or preventing translation or splicing.
A new type of antisense RNA, splice-switching oligonucleotides (SSOs), can modify gene expression by fixing abnormal splicing, unlike siRNAs, which suppress protein expression. However, it is often difficult to evaluate the effectiveness of SSOs.
The capacity of ASOs to interact with pre-mRNA allows them to target splicing processes. In addition, it dramatically expands the number of RNA sequences that can be selected for ASO binding, reducing off-target effects. For example, only 7% of the 2842 known single-nucleotide polymorphisms in the HTT gene, which codes for the huntingtin protein, can be targeted in mature mRNA (using siRNAs). Still, PCR can target these single-nucleotide polymorphisms 100% of the time.
siRNA stands for small interfering RNA. Small interfering RNAs (siRNAs) are double-stranded RNA molecules that use the RNA-induced silencing complex (RISC) to silence genes. They are 19–23 base pairs long (with a two-nucleotide 3′ overhang). The RISC complex binds siRNA, which is then unraveled via ATP hydrolysis and guided by the enzyme “Slicer” to target mRNA breakdown based on complementary base pairing. As a therapeutic product, siRNA can be delivered through the eye or nose, enhancing bioavailability. However, because intravenous injections require significant amounts, around 20–30% of the total blood volume, targeted distribution to treat malignancies is difficult. In direct tissue/organ electroporation, conjugation to membrane-permeable peptides, and liposome packing in vivo, exogenous siRNAs persist for a few days (at most, a few weeks in non-dividing cells). However, they can use the RISC system to control gene expression by base-pairing to mRNA targets and promoting their destruction when they reach their target.
2.1. “Biosimilar” mRNA Products
mRNA products are classified as biological drugs, despite being synthetic products. However, the FDA has stated that an approved vaccine may not require extensive testing
[5]. This observation has led to the concept of modified biological application wherein fewer studies are required if a copy of an mRNA product is presented
[6][7].
2.2. Synthetic Messenger RNA
Synthetic mRNA has proven its safety and efficacy, and its efficacy offers many opportunities in therapeutics, gene therapy, or vaccine applications
[8][9][10]. Using mRNA for personalized and more specific targets introduces new therapeutic modalities; for example, in vitro synthesis based on bacteriophage RNA polymerases, such as T7 or SP6
[11], removes cell culturing or extracting proteins using complex purification methods and offers a scalable manufacturing system
[12]. The first commercial-scale cell-free GMP mRNA product was introduced in 2023
[13].
2.3. mRNA Construct Design
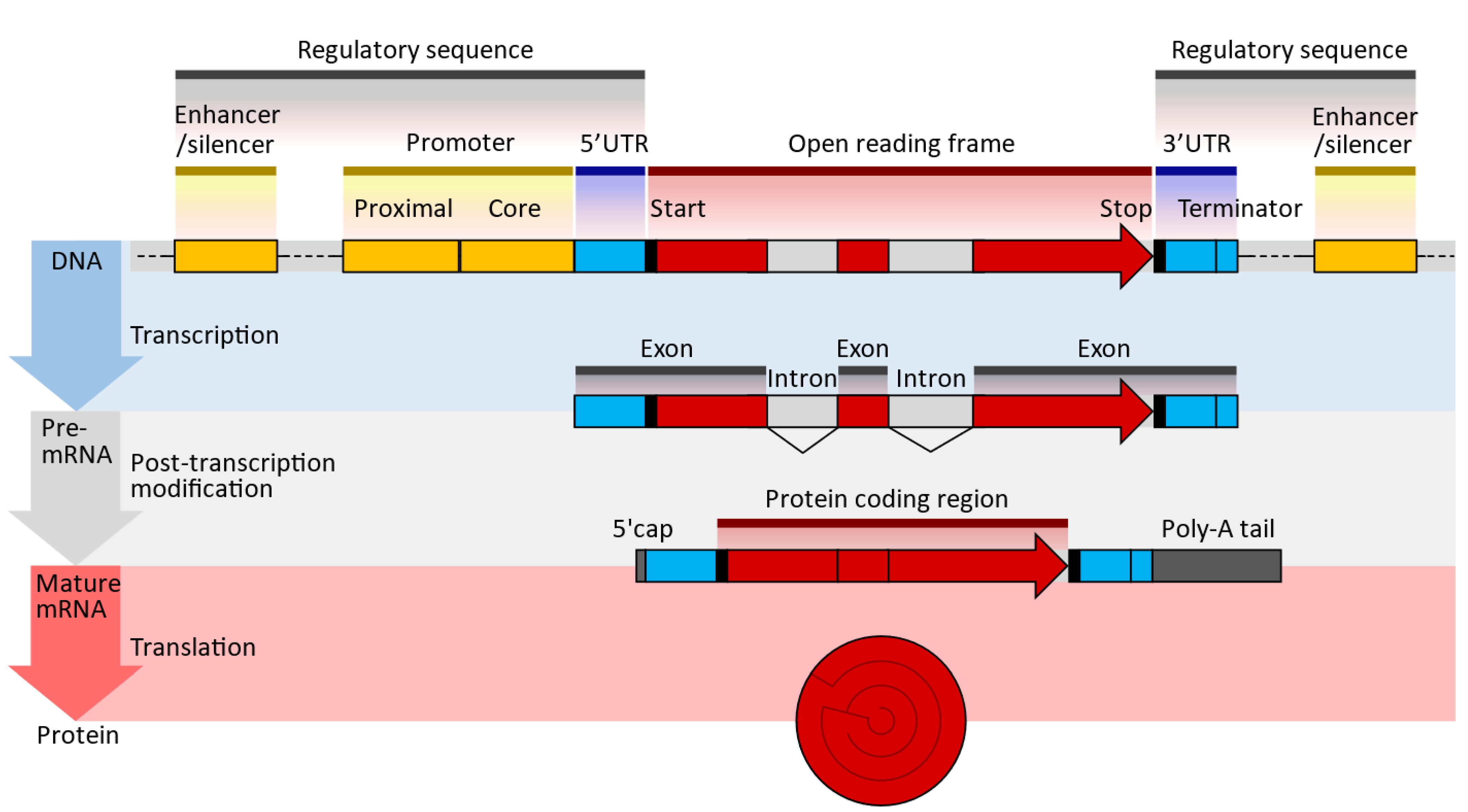
Typical DNA, pre-mRNA, and mRNA sequences are shown in
Figure 1 [14][15].
Figure 1. Typical DNA, pre-MRNA, and mRNA design sequences; UTR—untranslated region; poly(A)—polyadenylate signal tail (enumver as beeded
[16]). The 5′ cap at the end allows for sequence recognition, protecting the translated molecules from digestion by nucleases
[17]. The 5′UTR/3′UTR determines the translation efficiency, stability, and location; it is pivotal to optimizing expression
[16][18]. The open reading frame or coding sequence (CDS) lists the genes expressed. These genes are optimized and modified to improve translational efficiencies, such as the modification of guanine and cytosine content
[19]. The Poly(A) tail is essential for optimal translation 11 and improves stability by blocking digestion by 3′ exonuclease, increasing translation efficiency and adding to the molecule’s stability
[20].
The transcription of modified nucleic acids is another aspect of mRNA design. When introduced exogenously to a cell, mRNA can induce immunogenicity. However, the innate immune system is not activated when naturally occurring, chemically altered nucleosides, such as pseudouridine and 1-methyl pseudouridine, are present. Additionally, ribosomes translate nucleoside-modified mRNA more quickly than untreated mRNA
[21][22]. The sequences of the UTRs and the coding region are optimized to improve translation efficiency and mRNA stability. The UTRs of natural mRNAs, such as those of alpha- or beta-globin, are commonly used as the basis for the UTRs of mRNA vaccines. The 5′ cap of the mRNA strand is introduced either during in vitro transcription (IVT), by including a cap analog in the IVT reaction mixture, or after IVT, by using specialized enzymes (e.g., 2′-O-methyltransferase). Co-transcriptional capping gives lower yields than post-transcriptional capping but may be more cost-effective. Polyadenylation, or the addition of a poly(A) tail at the 3′ end of the mRNA strand, is performed enzymatically, since the poly(A) sequence is long, which will make the plasmid unstable. For example, while most tails are about 70–80 units long, the Pfizer COVID-19 vaccine has 110 tails.
2.4. Large-Scale mRNA Production
2.4.1. Template DNA Design and Preparation
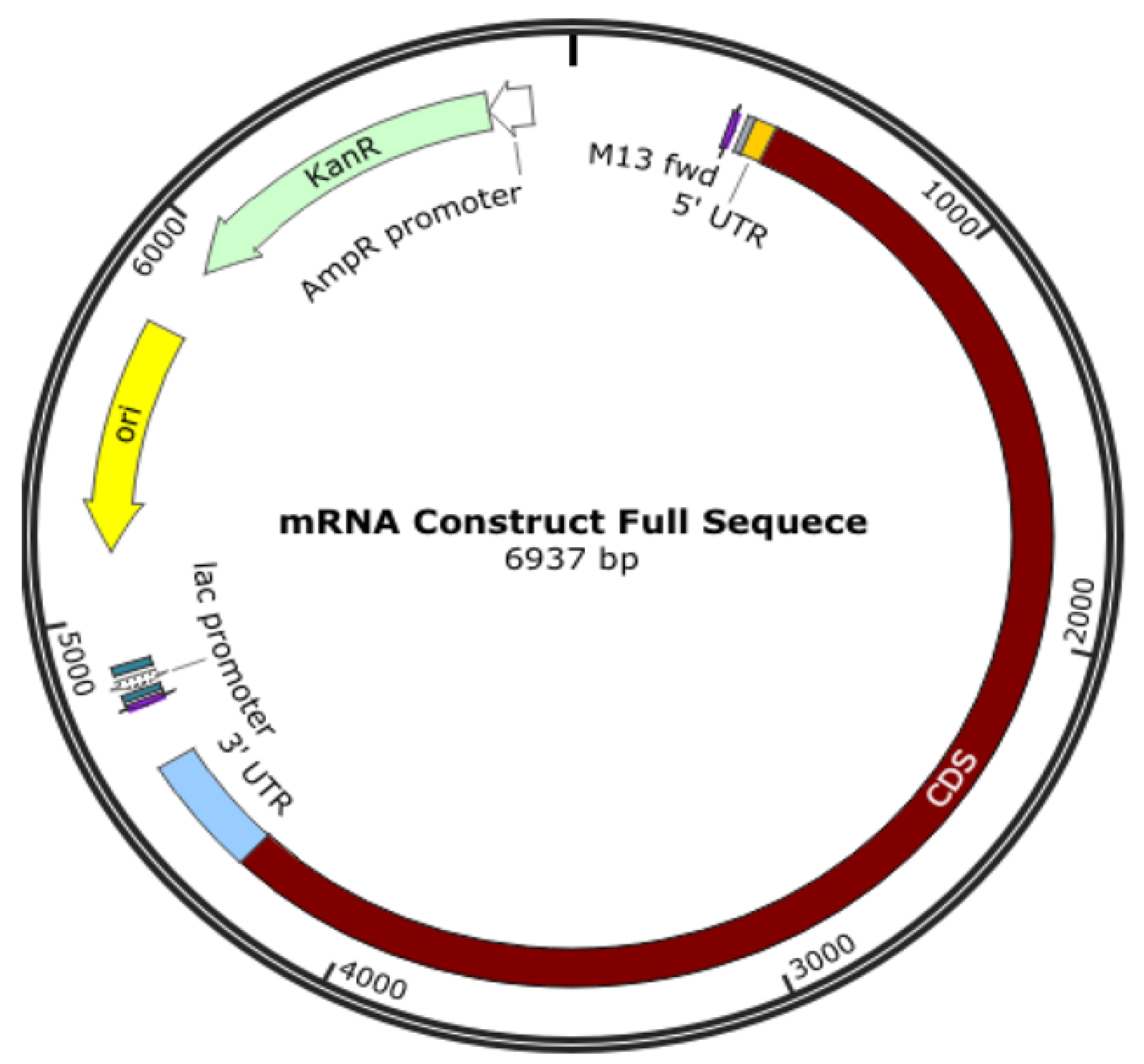
An oligonucleotide, a cDNA made from RNA, a plasmid construct, or a PCR output can all be used as the DNA template for mRNA in vitro transcription (IVT). The template has the desired sequence and a double-stranded promoter region where RNA polymerase (such as T7 or SP6) can attach to start the synthesis of RNA. Plasmid constructs are readily obtained from CROs, who can provide these quickly. mRNA production starts with the creation of DNA, which is split into RNA; the DNA comes from plasmid DNA, a circular DNA in E. coli. As shown in Figure 1, the plasmid DNA includes several modifications in the translating region; this generally involves nucleoside modification where uridine is replaced with pseudo-uridine, but this can also be carried out in the IVT process. However, if modifications to translated proteins are required, these modifications are made in the plasmid DNA. For example, the coronavirus’s surface protein sequence is modified to prevent it from collapsing (Figure 2).
Figure 2. Plasmid DNA is designed to produce mRNA for a COVID-19 vaccine.
The translating region may include nucleoside modification, such as replacing uridine with pseudo-uridine to reduce the immune response to exogenous mRNA and increases its stability to translate the specified sequence. It is also essential to know that the expressed protein need not be the same as the surface protein of a virus, and some modifications are made; for example, K986P and V987P for the COVID-19 vaccine are used to stabilize the translated protein to keep it from collapsing and being pushed out of the cell.
2.4.2. DNA Template Linearization
DNA linearization is a technique used to convert circular DNA molecules, such as plasmids or viral genomes, into linear DNA fragments. This process involves the cleavage of circular DNA at specific sites to generate linear DNA with defined ends. DNA linearization has various applications in molecular biology research, including DNA sequencing, cloning, gene editing, and gene expression studies.
There are several methods that are commonly used for DNA linearization:
-
Restriction enzyme digestion: Restriction enzymes, also known as restriction endonucleases, such as XbaI, cleave DNA at specific recognition sites. Selecting a restriction enzyme that recognizes a site within the circular DNA molecule makes it possible to generate linear DNA fragments with defined ends. The choice of restriction enzyme depends on the recognition site sequence and the desired DNA fragment size.
-
PCR amplification with primers containing restriction sites: PCR (polymerase chain reaction) can amplify a specific region of the circular DNA using primers containing restriction sites. The resulting PCR product can be digested with the corresponding restriction enzyme to linearize the DNA at the desired site.
-
CRISPR-Cas9 cleavage: CRISPR-Cas9 is a powerful gene editing tool that can be used to cleave DNA at specific target sites. By designing guide RNAs (sgRNAs) that target specific sites on the circular DNA, Cas9 nuclease can create double-stranded breaks, resulting in linear DNA fragments when repaired via cellular DNA repair mechanisms.
-
Chemical cleavage: Certain chemicals, such as hydroxylamine or osmium tetroxide, can cleave DNA at specific sites, resulting in linear DNA fragments. Chemical cleavage methods are less commonly used than restriction enzyme digestion or CRISPR-Cas9 cleavage, but can be useful in specific situations.
After linearization, the resulting DNA fragments can be purified and verified using gel electrophoresis or DNA sequencing methods. Linearized DNA can be used in various downstream applications, such as cloning into other vectors, DNA sequencing, gene expression studies, or gene editing using techniques such as CRISPR-Cas9.
It’s important to note that the choice of linearization method depends on the specific experimental requirements, such as the desired DNA fragment size, specific recognition sites, and the availability of appropriate enzymes or reagents. Nevertheless, the proper linearization of circular DNA is critical in many molecular biology experiments, and should be carefully optimized to ensure accurate and reproducible results
[23].
The 5′ cap, 3′ cap, and poly(A) tail are added during the IVT process or after transcription using capping enzymes and poly(A) polymerase
[24][25][26].
Most aspects of using chemically modified nucleosides or optimizing the nucleoside composition mix have been shown to reduce dsRNA byproducts more than threefold.
mRNA purification:
Unwanted side products from mRNA in vitro synthesis can include double-stranded (dsRNA), uncapped mRNA, and mRNA fragments in varying proportions. These byproducts may overestimate the total amount of functional mRNA, trigger innate immunity, or drastically impede mRNA translation. For size purification, high-performance liquid chromatography (HPLC) is commonly used. Clinical-grade mRNA for therapeutic applications must be free of impurities from upstream processes.
2.5. Final Formulation
Formulating lipid nanoparticle (LNP) mRNA products is a critical step in developing mRNA-based therapeutics, including mRNA vaccines and gene therapies. LNPs are lipid-based nanoparticles that encapsulate mRNA molecules, protect them from degradation, facilitate cellular uptake, and enable efficient mRNA delivery into cells. The formulation of LNP mRNA products involves several key components and steps.
Lipid selection: The choice of lipids is a crucial factor in LNP formulation. Different lipids with varying properties, such as cationic, ionizable, and PEGylated lipids, can form LNPs with distinct characteristics. For example, cationic lipids provide a positive charge to LNPs, enhancing cellular uptake, while ionizable lipids allow for the efficient release of mRNA within cells. In addition, PEGylated lipids can improve the stability and pharmacokinetics of LNPs.
mRNA encapsulation: The mRNA molecule carries the genetic information to be delivered into cells and is encapsulated within LNPs. mRNA is complexed with the lipids through electrostatic interactions, forming a lipid–mRNA complex that protects the mRNA from enzymatic degradation in the extracellular environment.
Formulation optimization: The formulation of LNPs is optimized to achieve desired characteristics, such as particle size, charge, and stability. Particle size is an important parameter that affects cellular uptake, biodistribution, and immune response. The surface charge of LNPs can also be controlled by adjusting the composition and ratio of lipids, which can influence their cellular uptake and intracellular trafficking.
Sterilization and quality control: To ensure product safety, LNPs intended for clinical use must be manufactured under sterile conditions. Sterilization methods, such as filtration or gamma irradiation, may be used to eliminate potential contaminants. In addition, quality control tests, including particle size analysis, encapsulation efficiency, and endotoxin testing, are performed to ensure the quality and consistency of LNP mRNA products.
Storage and stability: LNPs are typically stored at low temperatures, such as −20 °C or −80 °C, to maintain stability and prevent degradation. The stability of LNPs can also be influenced by factors such as lipid composition, pH, and storage conditions, and formulation optimization is necessary to ensure the long-term stability and shelf-life of the LNP mRNA products.
Characterization and validation: LNPs are characterized and validated to ensure their quality, safety, and efficacy. Various analytical techniques, such as dynamic light scattering (DLS), transmission electron microscopy (TEM), and reverse-phase high-performance liquid chromatography (RP-HPLC), may be used to characterize the size, morphology, and encapsulation efficiency of LNPs. In vitro and in vivo studies are also conducted to validate the biological activity and therapeutic potential of LNP mRNA products.
The formulation of LNP mRNA products is a complex and crucial process in developing mRNA-based therapeutics. Optimizing lipid composition, encapsulation efficiency, particle size, and stability is essential to ensuring the efficient mRNA delivery, cellular uptake, and therapeutic efficacy of LNP mRNA products. Rigorous quality control and characterization are also critical to ensuring the safety and effectiveness of LNP mRNA formulations for clinical use
[27].
2.6. Testing
As a new product class, the testing of RNA therapeutic products is still evolving. However, the tests associated with safety and delivery remain the same as those recommended for mRNA vaccines. The products should be tested for their nanoparticle profiles, encapsulation efficiency, in vitro toxicity due to formulation components, stability, storage condition, and shelf-life confirmation. In addition, cell-based assays can be used to test immune responses.
Serological analyses involve protein concentration and activity. Titration assays indirectly measure potency.
A major advance in the validation of RNA-based products has come from the US Pharmacopoeia; though it applies to mRNA vaccines, most of the proposed methods apply to all types of RNA products. In addition, using a USP method reduces the cost of method validation, as verification is only required for the USP tests
[28].