Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | ABHISHEK SHARMA | -- | 2475 | 2023-04-27 12:41:35 | | | |
| 2 | Dean Liu | Meta information modification | 2475 | 2023-05-04 02:54:30 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Majumdar, A.; Sharma, A.; Belludi, R. Antiviral Defence Mechanisms in Plants. Encyclopedia. Available online: https://encyclopedia.pub/entry/43572 (accessed on 08 February 2026).
Majumdar A, Sharma A, Belludi R. Antiviral Defence Mechanisms in Plants. Encyclopedia. Available at: https://encyclopedia.pub/entry/43572. Accessed February 08, 2026.
Majumdar, Anik, Abhishek Sharma, Rakesh Belludi. "Antiviral Defence Mechanisms in Plants" Encyclopedia, https://encyclopedia.pub/entry/43572 (accessed February 08, 2026).
Majumdar, A., Sharma, A., & Belludi, R. (2023, April 27). Antiviral Defence Mechanisms in Plants. In Encyclopedia. https://encyclopedia.pub/entry/43572
Majumdar, Anik, et al. "Antiviral Defence Mechanisms in Plants." Encyclopedia. Web. 27 April, 2023.
Copy Citation
Plant viruses, as obligate intracellular parasites, rely exclusively on host machinery to complete their life cycle. Whether a virus is pathogenic or not depends on the balance between the mechanisms used by both plants and viruses during the intense encounter. Antiviral defence mechanisms in plants can be of two types, i.e., natural resistance and engineered resistance. Innate immunity, RNA silencing, translational repression, autophagy-mediated degradation, and resistance to virus movement are the possible natural defence mechanisms against viruses in plants, whereas engineered resistance includes pathogen-derived resistance along with gene editing technologies.
Plant viruses
natural resistance
engineered resistance
1. Natural Resistance to Plant Viruses
1.1. Innate Antiviral Immunity
There are two layers of plant immune responses against microbial pathogens, i.e., PAMP (pathogen-associated molecular pattern) triggered immunity (PTI) and Effector-triggered immunity (ETI) (Figure 1 (1)). PTI is the initial mechanism by which plants detect the microbes at the cell membrane through detection of conserved PAMPs by extracellular pattern recognition receptors (PRRs) [1]. PRRs begin to dimerize as soon as the PAMPs are detected and associate with cofactors such as somatic embryogenesis receptor-like kinases (SERKs). This sets off a cascade of intracellular signalling events, such as the generation of reactive oxygen species (ROS), ion influx, increase in the production of defence hormones, and mitogen-activated protein kinases (MAPKs) activation. All these events result in the expression of the pathogenesis-related (PR) proteins, synthesis and deposition of callose at the plasmodesmata, and cell wall strengthening, leading to the generation of resistance response. [2]. Activation of PTI sometimes leads to hypersensitive response (HR) causing programmed cell death (PCD) that causes necrotic spots at the infection site [3]. Although plant PTI against other phytopathogens is well understood, plant viruses are traditionally known as non-PAMP coding pathogens [4]. However, some evidence implies that PTI also has a significant influence on both susceptible and resistant plant-virus interactions. For instance, exogenous application of double-strand RNA (dsRNA) caused SERK-1-dependent PTI responses in Arabidopsis [5]. In another instance, tobacco and Arabidopsis showed PTI-like responses due to the coat proteins of tobacco mosaic virus and potato virus X, respectively [6][7].
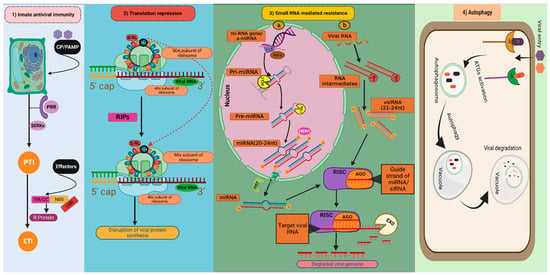
Figure 1. Natural antiviral defence mechanisms in plants: (1) Innate antiviral immunity; (2) Translation repression; (3) Small RNA-mediated resistance: (3a) MicroRNA mediated resistance. (3b) Small interfering RNA-mediated resistance. (4) Autophagy (Figure created in Biorender.com, accessed on 8 March 2023). CP/PAMP: Coat protein of virus/Pathogen-associated molecular patterns; PRR: Pattern recognition receptors; SERK: Somatic embryogenesis receptor kinases; PTI: PAMP triggered immunity; ETI: Effector-triggered immunity; S/RL: Sarcin/ricin loop; RIPs: Ribosome-inactivating proteins;  : depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.
: depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.
 : depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.
: depurination of S/R loop; miRNA gene/amiRNA: Micro RNA gene/artificial micro RNA; Pri-miRNA: Primary micro RNA; Pre-miRNA: Precursor micro RNA; miRNA: micro RNA; vsiRNA: Virus-derived small interfering RNA; DCL1: DICER LIKE 1; DCLs: DICER LIKE proteins; HST: HASTY; HEN1: HUA ENHANCER 1; RISK: RNA-induced silencing complex; AGO: Argonautes; EXO: Exonuclease; AR: Autophagy receptors; VF: Viral factors; ATGs: Autophagy-related genes.Pathogens in response to PTI introduce particular proteins called effectors in plant cells to weaken PTI-mediated defence. In response to these effectors, host plants depend on specific intracellular receptors known as R gene proteins which cause direct or indirect identification of the pathogen effector molecules. These R gene products can block the effectors and activate the effector-triggered immunity (ETI) [8]. R genes mediating resistance against various plant viruses have been extensively cloned over the last decade due to their apparent practical value. Functional R proteins are composed of three domains: a central nucleotide-binding site (NBS) domain, a leucine-rich repeat (LRR) domain, and an N terminal Toll Interleukin-1 receptor (TIR) or coiled-coil (CC) domain. Systemic acquired resistance (SAR) is a condition that can occur as a result of both PTI and ETI and is characterised by the development of resistance in the tissues that are distal to the infection [9]. While the non-expressor of PR1 (NPR1) is a protein with ankyrin domains, which is necessary for triggering salicylic acid (SA) signalling and establishing SAR, salicylic acid is the principal plant hormone responsible for establishing SAR [10]. SAR, together with pathogenesis-related proteins, confers resistance to the host plants against various pathogens [11].
1.2. Translation Repression as Virus Resistance
The initiation of translation is a critical step in protein synthesis that demands an array of eukaryotic initiation factors (eIFs). The translation of eukaryotic mRNA hinges on the association between the translation eukaryotic initiation factor 4E (eIF4E) and their 5′ m7 G cap structure, and the 3′ polyA tail’s interaction with the polyA-binding protein (PABP) also heightens the process. Although translation factors are not typically encoded by viruses, they have developed several techniques to hijack translation factors from their hosts, resulting in the promotion of viral RNA translation while compromising the translation of endogenous mRNAs. Mutated isoforms of the translation IFs, i.e., eIF4E and eIF4G have been found linked to many plant recessive resistance genes. These mutations typically prevent host factors from interacting with viral RNAs or proteins to suppress viral protein translation, an endogenous antiviral mechanism that has emerged recently [12]. Recessive resistance occurs when a component (or components) of the translation machinery recruited by viruses ceases to operate, resulting in a ‘loss of susceptibility’ to viruses. For instance, a family of proteins known as ribosome-inactivating proteins (RIPs) can inhibit the synthesis of new proteins by depurinating the sarcin/ricin loop (SRL) of rRNA [13] (Figure 1 (2)). The most thoroughly studied RIP with antiviral activity is the pokeweed antiviral protein (PAP) from Phytolacca americana. PAP slows down the spread of various plant viruses, including the cauliflower mosaic virus, potato virus X, and cucumber mosaic virus [14]. In the interaction between plants and viruses, small RNA-related translation suppression can be quite significant. For instance, in tomato plants infected with the tomato ringspot virus, recovery of symptoms is associated with the AGO1-dependent translation repression of viral RNA2. [15]. Additionally, the inability of one strain of plum pox virus to recruit translation initiation factors conferred resistance against this strain in wild-type Arabidopsis thaliana and Chenopodium foetidum [16].
1.3. Small RNA-Mediated Antiviral Defence
Small RNAs (sRNA) are essential for the epigenetic and post-transcriptional control of gene expression in plants throughout growth, developmental, and biotic/abiotic stress responses. Recent research has highlighted the role of two distinct families of small RNAs, known as small interfering RNA (siRNA) and micro RNA (miRNA) with respect to biotic stress responses in plants. By regulating the gene expression of the modulators of host defence pathways, these sRNAs activate antiviral defence during virus infection. RNA silencing, commonly referred to as RNA interference (RNAi), is a conserved evolutionary method for regulating endogenous expressions of genes and preventing the entry of foreign nucleic acids such as viruses and transposons [17]. The plant type III endoribonucleases or dicer-like (DCL) proteins detect and cleave the virus-derived double-stranded RNA (dsRNA) into small 20–24 nucleotide RNA duplexes known as virus-derived short interfering small RNAs (vsiRNAs) [18]. The vsiRNAs incorporated into argonaute proteins (AGOs) form the core component of the RNA-induced silencing complex (RISC), which cleaves homologous viral RNAs and/or suppresses translation of viral protein synthesis [19][20] (Figure 1 (3b))
Unlike siRNAs, miRNAs are endogenously produced non-coding short RNAs by RNA polymerase II from ssRNA precursors with a hairpin structure. [21]. Along with the well-established function of vsiRNAs, research findings suggest that miRNAs play a significant role in plant antiviral defence. At the initiation of miRNA synthesis, a primary miRNA transcript, or Pri-miRNA is synthesised after transcription and is made up of an incomplete stem-like structure of 100–120 nucleotides. Then, the precursor miRNA (Pre-miRNA) which is about 70 nucleotides long, formed after processing of Pri-mRNA by the DCL1 complex in the nucleus. Finally, the cleaving of 20–24 nucleotides from the initial cleavage point results in the formation and release of the miRNA/miRNA duplex from the stem-like structure. HUA Enhancer 1 (HEN1) methylates miRNA-miRNA duplex to protect it from degradation in the cytosol. Finally, they are transported to the cytosol by exportin-5 homolog HASTY (HST). In the cytosol, mature miRNAs are incorporated into RISC complexes having AGO proteins. The majority of the animal miRNAs that have been studied so far have miRNA and mRNA base pairing, which hinders the target mRNA translation. This is one of two ways that miRNAs guide RISC to down-regulate target mRNAs. Contrarily, the majority of plant miRNAs form substantial base pairing and directly cleave their target mRNAs. [22] (Figure 1 (3a)). Recently, the roles of various miRNAs, such as miR168, miR528, miR319, and miR444 in rice antiviral immunity have been identified. By modifying jasmonic acid (JA) signalling, MiR319 has been reported to confer antiviral resistance to rice against the rice-ragged stunt virus and to wheat against the rice black-streaked dwarf virus. [23].
1.4. Dominant Viral Resistance Genes
A few dominant resistance genes that function independently of the traditional innate immune signalling pathway have also been discovered in the past ten years. These dominant resistance genes express proteins that cannot be incorporated into the so-called plant innate immunity because they differ structurally from conventional R proteins. The majority of these dominant resistance genes inhibit the action of viral proteins by interacting with them in a direct manner. These dominant resistance gene products are referred to as atypical dominant viral resistance proteins (ADVRPs). The majority of ADVRPs that have been identified so far are from a protein family called lectin. Restricted TEV movement (RTM) 1, a member of the lectin-protein family is found in Arabidopsis, and it particularly provides resistance to a number of potyviruses, including TEV, lettuce mosaic virus (LMV), and plum pox virus by inhibiting their long-distance movement [24]. A lectin-like ADVRP of Arabidopsis called Jacaline-type lectin required for potexvirus resistance 1 (JAX1) imparts broad spectrum resistance to potexviruses at the initial infection stage by suppressing the activity of viral RNA-dependent RNA polymerase (RdRp) [25]. BanLec-1, another lectin from Musa paradisiaca, attaches to the TMV CP and inhibits virus infection in plants [26].
1.5. Resistance to Virus Movement within and between the Cells
For plant viruses to infect the entire plant system, they must spread from the initially infected cells to nearby ones. This must occur after the virus multiplication has begun within the cytoplasm and/or nucleus of a plant cell in a susceptible host. Host resistance to viral infection is evident when the virus only appears to affect one or a small number of cells but is unable to move past this initial focus of infection. At this point, resistance can be brought on by active host defence systems that swiftly thwart virus spread or by a breakdown in the connections between plant and viral components necessary for cell-to-cell movement. It is known that a variety of host gene alterations hinder the cell-to-cell movement of plant viruses. The cum1 and cum2 mutations in Arabidopsis cause reduced multiplication of CMV, thus rendering its movement in adjacent cells [27]. Pvr11 and Sbm1 were found to be mutated at a locus expressing eIF4E in pepper and pea, respectively [28][29]. eIF4E is thought to play a role in viral RNA replication or translation, although it may also be involved in cell-to-cell movement. The restricted-TEV-movement (RTM) genes RTM1, RTM2, and RTM3 prevent systemic tobacco etch virus (TEV) movement between plant cells. These genes interact with the virus CP and are expressed in phloem sieve elements [30]. Similar to this, BTR1 is a ribonucleoprotein K-homology RNA binding protein that binds to ToMV (tomato mosaic virus) genomic RNA and limits its movement between cells [31]. By inducing cell death at the sites of infection and restricting the movement between cells, the Ny-1 gene confers potato virus Y (PVY) resistance in potatoes [32].
1.6. Autophagy as Antiviral Mechanism against Plant Viruses
Proteins and defective organelles are transferred to vacuoles or lysosomes for destruction through autophagy, which is an evolutionarily conserved intracellular degradation mechanism [33]. Autophagy also contributes to antiviral defence mechanisms in plants. Virus particles upon entry are recognised by specific autophagy receptors (AR). After recognition, autophagy-related genes (ATGs) get activated and initiate the formation of autophagosome, which ultimately fuse with the vacuole leading to the vacuolar degradation of autophagosome contents which also harbour viral particles (Figure 1 (4)). As TMV aggregates at infection sites in Beclin-1 or ATG7 silenced plants, it has been hypothesised that autophagy contributes to plant defence against viruses. It is interesting to note that induction of autophagy extends beyond the TMV infection sites to uninfected neighbouring tissues, where it prevents cell death [34]. Several reports have recently emphasised the significance of this mechanism in restricting virus multiplication. To prevent the replication of the cotton leaf curl Multan virus (CLCuMuV), ATG8 specifically interacts with the beta-satellite (βC1) of the CLCuMuV [35]. Not only DNA viruses but RNA viruses are also targeted by the antiviral effects of autophagy. Nuclear inclusion protein B (NIb) of turnip mosaic virus (TuMV), interacts with autophagy-related gene 6 (ATG6, also known as Beclin1) to prevent viral replication [36]. Additionally, it was reported that TuMV infection triggered autophagy, which reduced viral RNA accumulation in Arabidopsis. [37].
1.7. Cross Protection
Cross protection has been utilised to manage viral diseases together with protein and RNA-mediated resistance. By infecting the host plant with a mild strain of the virus, the resistance is conferred effectively. The infection caused by attenuated strain makes the affected plant immune to further infections by a viral strain that is closely related to the inoculated virus. After the attenuated strain has been inoculated, it may trigger strain or sequence-specific resistance against the challenger virus. In a number of experiments, the effect of coat protein on cross-protection reactions has been examined. When systemically expressed using PVX as a virus vector, various TMV CP mutants with altered CP aggregation demonstrated that the CP mutants with high assembly capacity offered effective cross protection against TMV infections. TMV cross protection and CP-mediated resistance may therefore depend on CP’s capacity to prevent the challenging virus from uncoating its capsid [38]. Numerous researchers have also discovered that mutations to virus silencing suppressors can reduce the severity of symptoms. For instance, mutations in the 126 kD replicase, a suppressor of silencing from the pepper mild mottle virus (PMMoV), resulted in milder symptoms as well as offering pepper plants resistance through cross protection [39].
2. Engineered Resistance to Plant Viruses
Engineered resistance against plant viruses can be divided into two parts: pathogen-derived resistance (PDR) and gene editing technologies.
2.1. Pathogen-Derived Resistance
The majority of the functions played by plant viruses during the replication cycle are well-characterised since they have short genomes and few genes. Viruses are suitable targets for developing artificial resistance based on the concept of pathogen-derived resistance (PDR). Sanford and Johnston were the ones who originally introduced this idea [40]. The most popular and effective method of obtaining PDR has been the exploitation of virus CP genes. High levels of resistance in transgenic plants have been shown to result from host plants expressing the CP of a number of RNA viruses including TMV, PVX, CMV, and TRV. This suggests that CP-mediated resistance is dependent on inhibiting the disassembly of the infecting virus. A noteworthy achievement is the use of transgenic papaya expressing the CP transgenes of papaya ring spot virus for the control of papaya ringspot disease in Hawaii [41].
2.2. Gene Editing Technologies
These are further divided into Engineering ZFN or TALEN-based resistance and CRISPR/Cas technology.
References
- Schwessinger, B.; Zipfel, C. News from the frontline: Recent insights into PAMP-triggered immunity in plants. Curr. Opin. Plant Biol. 2008, 11, 389–395.
- Tang, D.; Wang, G.; Zhou, J.M. Receptor kinases in plant-pathogen interactions: More than pattern recognition. Plant Cell 2017, 29, 618–637.
- Coll, N.S.; Epple, P.; Dangl, J.L. Programmed cell death in the plant immune system. Cell Death Differ. 2011, 18, 1247–1256.
- Mandadi, K.K.; Scholthof, K.B.G. Plant immune responses against viruses: How does a virus cause disease? Plant Cell 2013, 25, 1489–1505.
- Alexopoulou, L.; Holt, A.C.; Medzhitov, R.; Flavell, R. Recognition of double-stranded RNA and activation of NF-κB by Toll-like receptor 3. Nature 2001, 413, 732–738.
- Allan, A.C.; Lapidot, M.; Culver, J.N.; Fluhr, R. An early tobacco mosaic virus-induced oxidative burst in tobacco indicates extracellular perception of the virus coat protein. Plant Physiol. 2001, 126, 97–108.
- Perraki, A.; Gronnier, J.; Gouguet, P.; Boudsocq, M.; Deroubaix, A.F.; Simon, V.; Germain, V. REM1. 3’s phospho-status defines its plasma membrane nanodomain organization and activity in restricting PVX cell-to-cell movement. PLoS Pathog. 2018, 14, e1007378.
- Dodds, P.N.; Rathjen, J.P. Plant immunity: Towards an integrated view of plant–pathogen interactions. Nat. Rev. Genet. 2010, 11, 539–548.
- Fu, Z.Q.; Dong, X. Systemic acquired resistance: Turning local infection into global defense. Annu. Rev. Plant Biol. 2013, 64, 839–863.
- Luna, E.; Bruce, T.J.; Roberts, M.R.; Flors, V.; Ton, J. Next-generation systemic acquired resistance. Plant Physiol. 2012, 158, 844–853.
- Kumar, D. Plant immune response strategies against pathogens. Plant Arch. 2020, 20, 1169–1174.
- Sanfaçon, H. Plant translation factors and virus resistance. Viruses 2015, 7, 3392–3419.
- Truniger, V.; Aranda, M.A. Recessive resistance to plant viruses. Adv. Virus Res. 2009, 75, 119–231.
- Domashevskiy, A.V.; Cheng, S.Y. Translation Initiation Complex eIFiso4F Targets Pokeweed Antiviral Protein (PAP) to Selectively Depurinate Uncapped Tobacco Etch Virus (TEV) RNA. Biophys. J. 2018, 114, 442.
- Pantaleo, V.; Szittya, G.; Burgyán, J. Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. J. Virol. 2007, 81, 3797–3806.
- Calvo, M.; Martínez-Turiño, S.; García, J.A. Resistance to Plum pox virus strain C in Arabidopsis thaliana and Chenopodium foetidum involves genome-linked viral protein and other viral determinants and might depend on compatibility with host translation initiation factors. Mol. Plant-Microbe Interact. 2014, 27, 1291–1301.
- Ding, S.W.; Han, Q.; Wang, J.; Li, W.X. Antiviral RNA interference in mammals. Curr. Opin. Immunol. 2018, 54, 109–114.
- Baulcombe, D. RNA silencing in plants. Nature. 2004, 431, 356–363.
- Llave, C. Virus-derived small interfering RNAs at the core of plant–virus interactions. Trends Plant Sci. 2010, 15, 701–707.
- Ghoshal, B.; Sanfaçon, H. Temperature-dependent symptom recovery in Nicotiana benthamiana plants infected with tomato ringspot virus is associated with reduced translation of viral RNA2 and requires ARGONAUTE 1. Virology 2014, 456, 188–197.
- Yu, Y.; Jia, T.; Chen, X. The ‘how’and ‘where’of plant micro RNAs. New Phytol. 2017, 216, 1002–1017.
- Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53.
- Zhang, C.; Ding, Z.; Wu, K.; Yang, L.; Li, Y.; Yang, Z.; Wu, J. Suppression of jasmonic acid-mediated defense by viral-inducible microRNA319 facilitates virus infection in rice. Mol. Plant. 2016, 9, 1302–1314.
- Ranf, S.; Gisch, N.; Schäffer, M.; Illig, T.; Westphal, L.; Knirel, Y.A.; Scheel, D. A lectin S-domain receptor kinase mediates lipopolysaccharide sensing in Arabidopsis thaliana. Nat. Immunol. 2015, 16, 426–433.
- Sugawara, K.; Shiraishi, T.; Yoshida, T.; Fujita, N.; Netsu, O.; Yamaji, Y.; Namba, S. A replicase of potato virus X acts as the resistance-breaking determinant for JAX1-mediated resistance. Mol. Plant-Microbe Interact. 2013, 26, 1106–1112.
- Yoshida, T.; Shiraishi, T.; Hagiwara-Komoda, Y.; Komatsu, K.; Maejima, K.; Okano, Y.; Namba, S. The plant noncanonical antiviral resistance protein JAX1 inhibits potexviral replication by targeting the viral RNA-dependent RNA polymerase. J. Virol. 2019, 93, e01506-18.
- Yoshii, M.; Yoshioka, N.; Ishikawa, M.; Naito, S. Isolation of an Arabidopsis thaliana mutant in which accumulation of cucumber mosaic virus coat protein is delayed. Plant J. 1998, 13, 211–219.
- Gao, Z.; Johansen, E.; Eyers, S.; Thomas, C.L.; Noel Ellis, T.H.; Maule, A.J. The potyvirus recessive resistance gene, sbm1, identifies a novel role for translation initiation factor eIF4E in cell-to-cell trafficking. Plant J. 2004, 40, 376–385.
- Ruffel, S.; Dussault, M.H.; Palloix, A.; Moury, B.; Bendahmane, A.; Robaglia, C.; Caranta, C. A natural recessive resistance gene against potato virus Y in pepper corresponds to the eukaryotic initiation factor 4E (eIF4E). Plant J. 2002, 32, 1067–1075.
- Chisholm, S.T.; Parra, M.A.; Anderberg, R.J.; Carrington, J.C. Arabidopsis RTM1 and RTM2 genes function in phloem to restrict long-distance movement of tobacco etch virus. Plant Physiol. 2001, 127, 1667–1675.
- Fujisaki, K.; Ishikawa, M. Identification of an Arabidopsis thaliana protein that binds to tomato mosaic virus genomic RNA and inhibits its multiplication. Virology 2008, 380, 402–411.
- Lukan, T.; Baebler, Š.; Pompe-Novak, M.; Guček, K.; Zagorščak, M.; Coll, A.; Gruden, K. Cell death is not sufficient for the restriction of potato virus Y spread in hypersensitive response-conferred resistance in potato. Front. Plant Sci. 2018, 9, 168.
- Klionsky, D.J.; Codogno, P. The mechanism and physiological function of macroautophagy. J. Innate Immun. 2013, 5, 427–433.
- Espert, L.; Codogno, P.; Biard-Piechaczyk, M. Involvement of autophagy in viral infections: Antiviral function and subversion by viruses. J. Mol. Med. 2007, 85, 811–823.
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Liu, Y. Autophagy functions as an antiviral mechanism against geminiviruses in plants. eLife 2017, 6, e23897.
- Li, F.; Zhang, C.; Li, Y.; Wu, G.; Hou, X.; Zhou, X.; Wang, A. Beclin1 restricts RNA virus infection in plants through suppression and degradation of the viral polymerase. Nat. Commun. 2018, 9, 1268.
- Hafrén, A.; Üstün, S.; Hochmuth, A.; Svenning, S.; Johansen, T.; Hofius, D. Turnip mosaic virus counteracts selective autophagy of the viral silencing suppressor HCpro. Plant Physiol. 2018, 176, 649–662.
- Bazzini, A.A.; Asurmendi, S.; Hopp, H.E.; Beachy, R.N. Tobacco mosaic virus (TMV) and potato virus X (PVX) coat proteins confer heterologous interference to PVX and TMV infection, respectively. J. Gen. Virol. 2006, 87, 1005–1012.
- Yoon, J.Y.; Ahn, H.I.; Kim, M.; Tsuda, S.; Ryu, K.H. Pepper mild mottle virus pathogenicity determinants and cross protection effect of attenuated mutants in pepper. Virus Res. 2006, 118, 23–30.
- Sanford, J.C.; Johnston, S.A. The concept of parasite-derived resistance-deriving resistance genes from the parasite’s own genome. J. Theor. Biol. 1985, 113, 395–405.
- Ferreira, S.A.; Pitz, K.Y.; Manshardt, R.; Zee, F.; Fitch, M.; Gonsalves, D.E. Virus coat protein transgenic papaya provides practical control of papaya ringspot virus in Hawaii. Plant Dis. 2002, 86, 101–105.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
836
Revisions:
2 times
(View History)
Update Date:
04 May 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




