Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Miryam Palacios-Pérez | -- | 2170 | 2023-04-02 03:20:15 | | | |
| 2 | Rita Xu | Meta information modification | 2170 | 2023-04-03 04:12:07 | | | | |
| 3 | Rita Xu | Meta information modification | 2170 | 2023-04-03 04:14:29 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
López-Cortés, G.I.; Palacios-Pérez, M.; Hernández-Aguilar, M.M.; Veledíaz, H.F.; José, M.V. Human Coronavirus Cell Receptors. Encyclopedia. Available online: https://encyclopedia.pub/entry/42717 (accessed on 07 February 2026).
López-Cortés GI, Palacios-Pérez M, Hernández-Aguilar MM, Veledíaz HF, José MV. Human Coronavirus Cell Receptors. Encyclopedia. Available at: https://encyclopedia.pub/entry/42717. Accessed February 07, 2026.
López-Cortés, Georgina I., Miryam Palacios-Pérez, Margarita M. Hernández-Aguilar, Hannya F. Veledíaz, Marco V. José. "Human Coronavirus Cell Receptors" Encyclopedia, https://encyclopedia.pub/entry/42717 (accessed February 07, 2026).
López-Cortés, G.I., Palacios-Pérez, M., Hernández-Aguilar, M.M., Veledíaz, H.F., & José, M.V. (2023, April 02). Human Coronavirus Cell Receptors. In Encyclopedia. https://encyclopedia.pub/entry/42717
López-Cortés, Georgina I., et al. "Human Coronavirus Cell Receptors." Encyclopedia. Web. 02 April, 2023.
Copy Citation
Coronaviruses interact with protein or carbohydrate receptors through their spike proteins to infect cells. Even if the known protein receptors for these viruses have no evolutionary relationships, they do share ontological commonalities that the virus might leverage to exacerbate the pathophysiology. ANPEP/CD13, DPP IV/CD26, and ACE2 are the three protein receptors that are known to be exploited by several human coronaviruses. These receptors are moonlighting enzymes involved in several physiological processes such as digestion, metabolism, and blood pressure regulation; moreover, the three proteins are expressed in kidney, intestine, endothelium, and other tissues/cell types.
coronavirus receptor
ANPEP/ CD13
DPP-IV/ CD26
1. Introduction
The designation Coronaviridae refers to a family of viruses that all display protein homotrimers across the whole surface of their viral membrane, giving them a crown-like appearance. The coronaviruses are encapsulated positive-stranded RNA viruses [1], and the family contains four distinct genera (alpha, beta, gamma, and delta coronavirus) all of which encode nearly all functionally identical proteins [2]. Known human coronaviruses belong specifically to the genera Alphacoronavirus or Betacoronavirus, whose common ancestor infected bats [2]. Table 1 depicts the seven distinct coronaviruses that can infect humans: HCoV- 229E, HCoV- NL63, HCoV- OC43, HCoV- HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2. The latter three have been the main causes of epidemics in recent years [1][3][4].
Table 1. Human coronaviruses and their most probable ancestor host. Seven human coronaviruses have emerged from zoonotic transmission events.
| Coronavirus | Genera | Identified | Most Probable Ancestor Host | Receptor | Ref |
|---|---|---|---|---|---|
| HCoV-229 E | Alphacoronavirus | 1965 | Bats Hipposideros and camelids | ANPEP/CD13 | [1][5][6] |
| HCoV-OC43 | Betacoronavirus | 1967 | Rodents and swine | Sialic acid | [7] |
| SARS-CoV | Betacoronavirus | 2002 | Bat Rhinolophus and civet | ACE2 | [8] |
| HCoV-NL63 | Alphacoronavirus | 2004 | Bats Triaenops | ACE2 | [6] |
| HCoV-HKU1 | Betacoronavirus | 2005 | Rodents | Sialic acid | [8][9] |
| MERS-CoV | Betacoronavirus | 2012 | Bat and camel | DPP IV/CD26 | [10] |
| SARS-CoV-2 | Betacoronavirus | 2019 | Bat Rhinolophus affini and | ACE2 | [11] |
The structural glycoprotein known as the spike protein (S) enables the host receptor identification and entry into the cell [12]. The S proteins of coronaviruses must be cleaved to activate the fusion peptide and infect the cell [13]. S proteins have two functional subdomains that allow membrane fusion. The host recognition involves the subdomain 1 (S1), which has a C-terminal domain (CTD) and an N-terminal domain (NTD). The receptor binding domain (RBD) may be in the NTD or the CTD, depending on the virus. The fusion machinery, which consists of the fusion peptide and the heptad repeats required for the fusion, is present in the second subdomain (S2). The S2 sequence is far more conserved among the four genera than the S1 sequence, as the virus must adapt to the receptors of the hosts [14]. The adaptability of the coronaviruses has led to their evolutionary success because they respond rapidly to selective pressures [15][16] and can jump to another species with highly identical receptors [14][17].
Coronaviruses are known to primarily use a host receptor, which can be either a protein or a carbohydrate [17]. It is hypothesized that an ancient coronavirus acquired a host galectin sequence that resulted in the ability to bind carbohydrates, so binding to carbohydrates is a more recent feature [18]. Even though each coronavirus has a primary host receptor, it has been shown that several S proteins can bind to other components of the cell membrane [19][20][21]. Even more, the S protein’s glycosylation can also interact with the lectins of the host [20]. The reported protein receptors employed by mammalian coronaviruses are the murine carcinoembryonic antigen-related cell adhesion molecule 1a (mCEACAM 1a), aminopeptidase N (ANPEP also CD13), dipeptidyl protease IV (DPP- IV also CD26), and angiotensin converting enzyme 2 (ACE2) [17]. Since the first one is receptor for the murine hepatitis virus (MHV) and it does not infect humans, herein, researchers will discuss the other three enzymes solely.
2. Human Protein Receptors for Coronaviruses Are Proteases
The viral receptors ANPEP, DPP- IV, and ACE2 display functional similarities, starting with the fact that they are all proteases, expressed in the membrane as dimers, and able to be shed from the membrane while still being enzymatically active. These proteases are moonlighting enzymes that participate in numerous processes and are expressed in a wide variety of cell types, and they are regarded as receptors due to their participation in signal-transduction pathways. In Figure 1A–E, the three proteases are shown in association with the coronavirus S proteins’ RBD region.
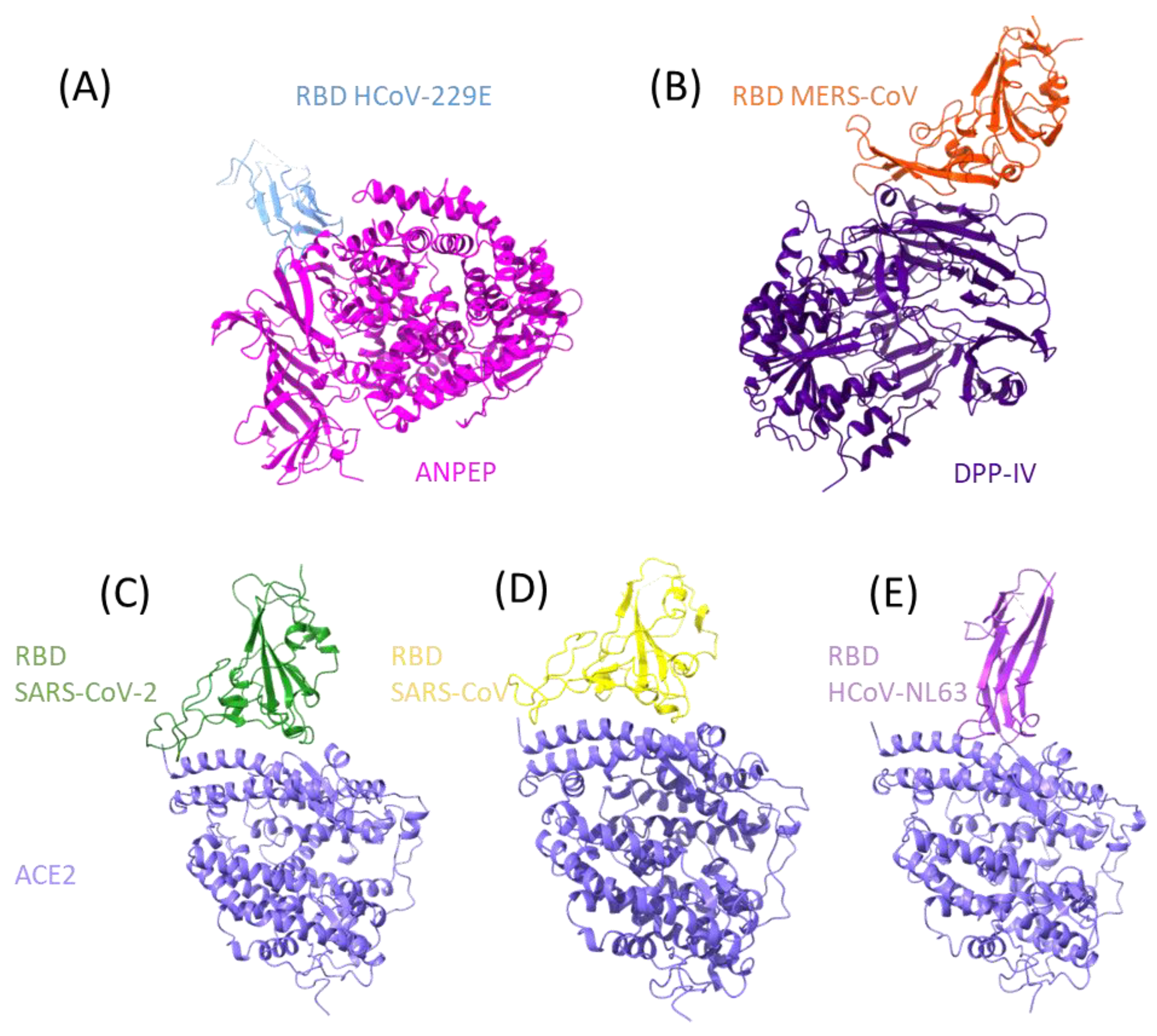
Figure 1. RBD interactions of human coronaviruses with their human protein receptor. The structures of protein–protein interaction of human coronaviruses to the human receptor is presented. At the top from left to right the RBD of HCoV-229E ((A), PDB: 6ATK) with ANPEP and the RBD of MERS-CoV ((B), PDB: 4KR0) with DPP- IV, at the bottom the RBD of SARS2-S ((C), PDB: 6M0J), SARS-S ((D), PDB: 2AJF) and HCoV-NL63 ((E), PDB: 3KBH) interacting with ACE2.
The three proteases process several substrates sequentially in the same pathways. A variety of enzymes, including ACE2 and ANPEP, are involved in the processing of vasoactive peptides of the renin-angiotensin aldosterone system (RAAS). Angiotensin I and II are digested by ACE2, and angiotensin III is subsequently cleaved by ANPEP to create angiotensin IV, which is then degraded by the same protease. This cascade will be explained in detail latter. The chemokine CXCL11 is initially hydrolyzed by DPP- IV and subsequently by ANPEP [22]. Other substrates could be cleaved by various proteases in order to be active, such as enkephalin, which is cleaved by ANPEP and subsequently by CD10 [23]. Additionally, potential substrates might be involved in other processes, such as digestion, sodium regulation, and blood-pressure regulation, where the proteases are co-expressed in epithelial cells of the brush border of the intestine, the renal epithelial cells and endothelial cells.
2.1. ANPEP
Initially discovered for its catalytic activity, the aminopeptidase N (ANPEP, CD13, alanyl peptidase, gp150) (EC: 3.4.11.2) is a glycoprotein of approximately 150 kDa. It was later found to be the same protein as CD13, which was a myeloid leukemia marker [24]. Even though various names have been given to it (including MY7, SJ1D1, and L138), aminopeptidase N (ANPEP) and CD13 remain [25][26]. ANPEP is a zinc2+ dependent metalloproteinase and catalyzes the hydrolysis of peptide bonds in the N-terminal end of neutral residues. The catalytic mechanism of porcine ANPEP was characterized as follows: Glu406, His383, and His387 chelate the Zn2+ ion, turning a water molecule into a catalyst, the proton is then transferred to Glu384, and finally this proton is passed to the nitrogen group of the protein [27]. Due to their high level of similarity, mammalian aminopeptidases have a comparable catalytic mechanism. In addition to this property, different species host various coronaviruses using the homologue receptor, e.g., pig transmissible gastroenteritis virus (TGEV), feline enteric coronavirus (FCoV), and canine enteric coronavirus (CCoV) [28].
Numerous substrates indicate that ANPEP is involved in a variety of physiological activities. Vasopeptides such as angiotensin III (Ang III, angiotensin 2-8), angiotensin IV (Ang IV); neuropeptides such as enkephalins, endorphins, neurokinin A, and nociceptin; hormones such as kallidan and bradykinin, glutathione, thymopentin, and splenopentin; extracellular matrix proteins such as entactin and collagen type IV; and inflammatory mediators such as kinins, kallidin, tufstin, CXCL11, and IL-8; and hemorphins are some of the substrates of human ANPEP [22][29][30].
ANPEP is a type II transmembrane protein, meaning that its amino terminus is cytoplasmic and its carboxyl terminus is extracellular (Figure 1A). Human ANPEP is composed of 967 amino acids, with a single transmembrane junction, a sizable extracellular region, and a small cytoplasmic portion of only nine residues [22]. Carbohydrates add about 40% of weight to the 109.5 kDa of protein, bringing the total weight to 150 kDa [31].
Noteworthily, ANPEP is expressed in the membrane, where it creates persistent non-covalently coupled homodimers [27]. The expression of this peptidase is constitutive in epithelial cells of the intestine, kidney, glomeruli, endothelial cells, fibroblasts, some neurons, meninges, choroid plexus, pineal gland, paraventricular nucleus, pituitary gland, and myeloid cells. ANPEP has also found to be a marker of myeloid cell differentiation. Interestingly, many cancer cells overexpress this protease [22], and in fact ANPEP has been the subject of research as a potential anti-cancer therapeutic drug [32]. This enzyme can be found in the cell membrane, in vesicles and as a soluble protein.
Despite its nine cytoplasmic amino acids and that is a membrane enzyme, it can trigger signaling cascades [33][34][35][36][37], although it is considered as an orphan receptor as the natural ligand is still unknown. Either as an enzyme or as a receptor, ANPEP is involved in a variety of processes, including angiogenesis, peptide breakdown during digestion, migration, cell adhesion, aggregation, and phagocytosis [22][30][37][38][39][40][41].
2.2. DPP-IV
The enzyme dipeptidyl peptidase IV (DPP-IV, CD26, adenosine deaminase 2 (ADCP2), (EC: 3.4.14.5)) has a serine peptidase activity that cleaves dipeptides at the amino terminal end [26][42]. Among its substrates are protein 1 type glucagon (GLP-1), gastric inhibitory protein (GIP), and substance P [43][44]. DPP-IV is a type II transmembrane protein with only one transmembrane passage and six cytoplasmic amino acids, but most of the protein (738 amino acids) is extracellular [43]. Two functional domains have been characterized in the extracellular region; one domain is homologous to an α/β hydrolase with serine protease activity and the other is composed by two subdomains, a cysteine-rich region and a highly glycosylated region (Figure 1B). The structure of CD26 is stabilized by a total of five disulphide bridges and nine N-glycosylations. The glycosylation of DPP-IV eases the formation of dimers and the interaction with other proteins [45], and the glycosylation patterns affect its cellular localization without altering its functions [46].
Exopeptidase DPP-IV prefers proline in the second amino acid position, alanine in a lesser degree, and glycine with the least preference when breaking down peptides [46]. Ser630, Asp708, and His740 all contribute to the catalysis [47]. DPP-IV can be released from the cell as an extracellular soluble enzyme by cleavage of its transmembrane region.
A range of leukocyte populations, including T, B, NK, and dendritic cells, express DPP-IV, as well as fibroblast, endothelial cells, epithelial cells of the kidney, liver, lung, small intestine, esophagus, breast, and prostate [48]. In contrast to TNF-α, which inhibits the expression of the protease, cytokine IL-12 causes an overexpression of this enzyme in activated T lymphocytes [49].
DPP-IV is important in the activation and maturation of T lymphocytes as it interacts with different proteins involved in the lymphocyte signaling pathway [50][51][52][53][54]. It also interacts with the enzyme adenosine deaminase (ADA) [55], which in turn interacts with adenosine receptor 2 expressed on dendritic cells [50]. Consequently, when these three proteins interact, a bridge between T cells and DCs is created. DPP-IV is associated with cellular adhesion because it can also bind to fibronectin and type 1 collagen in the extracellular matrix, an ability that metastatic cancer cells exploit [42].
2.3. ACE2
Angiotensin converting enzyme 2 (ACE2, EC: 3.4.17.23) is an 805 amino acid carboxypeptidase involved in the RAAS that regulates blood pressure; this regulation is focused on managing both blood volume and the resistance of the vascular system. Because of their involvement in this crucial physiological process, the most studied substrates of ACE2 are angiotensin I and angiotensin II, which are hydrolyzed to angiotensin 1-9 and angiotensin 1-7, respectively. Angiotensin 1-7 is a vasodilator, inhibits proliferation, and counteracts the vasoconstrictor effects of angiotensin II [56]. Besides this, ACE2 also has other substrates, such as neurotensin, kinetensin, des-Arg- bradykinin, apelins, and casomorphins, among others [57].
ACE2 is a type I transmembrane protein (Figure 1C–E); most of its amino acids are found on the extracellular side, followed by a transmembrane passage, and 43 cytoplasmic amino acids at the carboxyl terminus. Unlike the aforementioned proteases, ACE2 does have canonical signaling motifs; for example, it has a LIR motif (LC3-interacting region), an SH2-binding motif, a PTB, and a PDZ-binding motif [58]. Additionally, residues Tyr781 and Ser783 in the cytoplasmic region are susceptible to phosphorylation [59]. In fact, these motifs interact with integrin β3 and clathrin adaptor AP2 µ2 to enhance endocytosis and with the autophagy receptor MAP1LC3 and GABARAP [59]. On the extracellular side, the C-terminal domain of the protease is homologous to collectrin. It has a binding site for zinc2+ and Cl− ions; and it has seven glycosylation sites and three disulphide bridges.
The expression of this protease is also widespread, including endothelial cells, enterocytes, and Leydig and Sertoli cells; in the renal proximal tubule, heart, testicles, and type II pneumocytes; epithelial cells of bronchi, nose, cornea, brain, liver, and gallbladder [60]; and in the basal epidermal layer of the skin [61]. The expression of ACE2 in the epithelial cells of the small intestine is crucial for the co-expression of the neutral amino acid transporter [61]. ACE2 may be found on the cell membranes, in vesicles, secreted, and in the cytoplasm. In fact, the metalloproteinase ADAM17, also known as the tumor necrosis factor-a-converting enzyme (TACE), cleaves the membrane ACE2 and releases it into the interstitial space [62]. In addition, ADAM17 catalyzes the conversion of pro-TNF-α to TNF-α, which can increase ACE2 expression [60]; this upregulation has also been observed in the case of exposure to tobacco smoke and the chronic presence of inflammatory cytokines such as IFN-α, IFN-γ, and IL-13 [63].
The functions that depend on ACE2’s enzymatic activity include the maturation of vasoactive peptides, fluid pressure homeostasis, promoting myocyte contraction, and regulating cell proliferation [64]. The enzymatic site is apart from the binding site from SARS-CoV and SARS-CoV-2, so infection does not perturbate the enzymatic activity. Nonetheless, after either of the two viruses’ entry, ACE2 is downregulated, causing alterations of the RAS.
References
- Cui, J.; Li, F.; Shi, Z.-L. Origin and evolution of pathogenic coronaviruses. Nat. Rev. Microbiol. 2019, 17, 181–192.
- Woo, P.C.Y.; Lau, S.K.P.; Lam, C.S.F.; Lau, C.C.Y.; Tsang, A.K.L.; Lau, J.H.N.; Bai, R.; Teng, J.L.L.; Tsang, C.C.C.; Wang, M.; et al. Discovery of Seven Novel Mammalian and Avian Coronaviruses in the Genus Deltacoronavirus Supports Bat Coronaviruses as the Gene Source of Alphacoronavirus and Betacoronavirus and Avian Coronaviruses as the Gene Source of Gammacoronavirus and Deltacoronavirus. J. Virol. 2012, 86, 3995–4008.
- Zhao, X.; Ding, Y.; Du, J.; Fan, Y. 2020 update on human coronaviruses: One health, one world. Med. Nov. Technol. Devices 2020, 8, 100043.
- Zhou, P.; Yang, X.-L.; Wang, X.-G.; Hu, B.; Zhang, L.; Zhang, W.; Si, H.-R.; Zhu, Y.; Li, B.; Huang, C.-L.; et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273.
- Corman, V.M.; Baldwin, H.J.; Tateno, A.F.; Zerbinati, R.M.; Annan, A.; Owusu, M.; Nkrumah, E.E.; Maganga, G.D.; Oppong, S.; Adu-Sarkodie, Y.; et al. Evidence for an Ancestral Association of Human Coronavirus 229E with Bats. J. Virol. 2015, 89, 11858–11870.
- Tao, Y.; Shi, M.; Chommanard, C.; Queen, K.; Zhang, J.; Markotter, W.; Kuzmin, I.V.; Holmes, E.C.; Tong, S. Surveillance of Bat Coronaviruses in Kenya Identifies Relatives of Human Coronaviruses NL63 and 229E and Their Recombination History. J. Virol. 2017, 91, e01953-16.
- Kapikian, A.Z. The coronaviruses. Dev. Biol. Stand 1975, 28, 42–64.
- Corman, V.M.; Muth, D.; Niemeyer, D.; Drosten, C. Hosts and Sources of Endemic Human Coronaviruses. Adv. Virus Res. 2018, 100, 163–188.
- van der Hoek, L. Human coronaviruses: What do they cause? Antivir. Ther. 2007, 12, 651–658.
- Corman, V.M.; Eckerle, I.; Bleicker, T.; Zaki, A.; Landt, O.; Eschbach-Bludau, M.; van Boheemen, S.; Gopal, R.; Ballhause, M.; Bestebroer, T.M.; et al. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Eurosurveillance 2012, 17, 20285.
- Singh, D.; Yi, S.V. On the origin and evolution of SARS-CoV-2. Exp. Mol. Med. 2021, 53, 537–547.
- Li, F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261.
- Millet, J.K.; Whittaker, G.R. Host cell proteases: Critical determinants of coronavirus tropism and pathogenesis. Virus Res. 2015, 202, 120–134.
- López-Cortés, G.I.; Palacios-Pérez, M.; Zamudio, G.S.; Veledíaz, H.F.; Ortega, E.; José, M.V. Neutral evolution test of the spike protein of SARS-CoV-2 and its implications in the binding to ACE2. Sci. Rep. 2021, 11, 18847.
- López-Cortés, G.I.; Palacios-Pérez, M.; Veledíaz, H.F.; Hernández-Aguilar, M.; López-Hernández, G.R.; Zamudio, G.S.; José, M.V. The Spike Protein of SARS-CoV-2 Is Adapting Because of Selective Pressures. Vaccines 2022, 10, 864.
- Jaimes, J.A.; André, N.M.; Chappie, J.S.; Millet, J.K.; Whittaker, G.R. Phylogenetic Analysis and Structural Modeling of SARS-CoV-2 Spike Protein Reveals an Evolutionary Distinct and Proteolytically Sensitive Activation Loop. J. Mol. Biol. 2020, 432, 3309–3325.
- Millet, J.K.; Jaimes, A.J.; Whittaker, G.R. Molecular diversity of coronavirus host cell entry receptors. FEMS Microbiol. Rev. 2020, 45, fuaa057.
- Peng, G.; Sun, D.; Rajashankar, K.R.; Qian, Z.; Holmes, K.V.; Li, F. Crystal structure of mouse coronavirus receptor-binding domain complexed with its murine receptor. Proc. Natl. Acad. Sci. USA 2011, 108, 10696–10701.
- Qi, F.; Qian, S.; Zhang, S.; Zhang, Z. Single cell RNA sequencing of 13 human tissues identify cell types and receptors of human coronaviruses. Biochem. Biophys. Res. Commun. 2020, 526, 135–140.
- Hoffmann, D.; Mereiter, S.; Jin Oh, Y.; Monteil, V.; Elder, E.; Zhu, R.; Canena, D.; Hain, L.; Laurent, E.; Grünwald-Gruber, C.; et al. Identification of lectin receptors for conserved SARS-CoV-2 glycosylation sites. bioRxiv 2021.
- Daly, J.L.; Simonetti, B.; Klein, K.; Chen, K.E.; Williamson, M.K.; Antón-Plágaro, C.; Shoemark, D.K.; Simón-Gracia, L.; Bauer, M.; Hollandi, R.; et al. Neuropilin-1 is a host factor for SARS-CoV-2 infection. Science 2020, 370, 861–865.
- Mina-Osorio, P. The moonlighting enzyme CD13: Old and new functions to target. Trends Mol. Med. 2008, 14, 361–371.
- Gabrilovac, J.; Čupić, B.; Breljak, D.; Zekušić, M.; Boranić, M. Expression of CD13/aminopeptidase N and CD10/neutral endopeptidase on cultured human keratinocytes. Immunol. Lett. 2004, 91, 39–47.
- Look, A.T.; A Ashmun, R.; Shapiro, L.H.; Peiper, S.C. Human myeloid plasma membrane glycoprotein CD13 (gp150) is identical to aminopeptidase N. J. Clin. Investig. 1989, 83, 1299–1307.
- Griffin, J.D.; Ritz, J.; Nadler, L.M.; Schlossman, S.F. Expression of myeloid differentiation antigens on normal and malignant myeloid cells. J. Clin. Investig. 1981, 68, 932–941.
- López-Cortés, G.I.; Díaz-Alvarez, L.; Ortega, E. Leukocyte Membrane Enzymes Play the Cell Adhesion Game. Front. Immunol. 2021, 12, 742292.
- Chen, L.; Lin, Y.-L.; Peng, G.; Li, F. Structural basis for multifunctional roles of mammalian aminopeptidase N. Proc. Natl. Acad. Sci. USA 2012, 109, 17966–17971.
- Tresnan, D.B.; Levis, R.; Holmes, K.V. Feline aminopeptidase N serves as a receptor for feline, canine, porcine, and human coronaviruses in serogroup I. J. Virol. 1996, 70, 8669.
- Danziger, R.S. Aminopeptidase N in arterial hypertension. Heart Fail. Rev. 2008, 13, 293–298.
- Lu, C.; Amin, M.A.; Fox, D.A. CD13/Aminopeptidase N Is a Potential Therapeutic Target for Inflammatory Disorders. J. Immunol. 2020, 204, 3–11.
- Luan, Y.; Xu, W. The Structure and Main Functions of Aminopeptidase N. Curr. Med. Chem. 2007, 14, 639–647.
- Garay-Canales, C.A.; Díaz-Alvarez, L.; Lopez-Cortes, G.I. Novel immunotherapy strategies involving matrix metalloproteinase (MMP) family. In Immunotherapy in Resistant Cancer: From the Lab Bench Work to Its Clinical Perspectives; Morales-Montor, J., Segovia-Mendoza, M., Eds.; Academic Press Inc.: Cambridge, MA, USA, 2020; Volume 2, pp. 227–247.
- Gabrilovac, J.; Breljak, D.; Čupić, B. Regulation of aminopeptidase N (EC 3.4.11.2; APN.; CD13) on the HL-60 cell line by TGF-β1. Int. Immunopharmacol. 2008, 8, 613–623.
- Santos, N.; Langner, J.; Herrmann, M.; Riemann, D. Aminopeptidase N/CD13 is directly linked to signal transduction pathways in monocytes. Eur. PMC 2000, 201, 22–32.
- Subramani, J.; Ghosh, M.; Rahman, M.M.; Caromile, L.A.; Gerber, C.; Rezaul, K.; Han, D.K.; Shapiro, L.H. Tyrosine Phosphorylation of CD13 Regulates Inflammatory Cell–Cell Adhesion and Monocyte Trafficking. J. Immunol. 2013, 191, 3905–3912.
- Ghosh, M.; Gerber, C.; Rahman, M.M.; Vernier, K.M.; Pereira, F.E.; Subramani, J.; Caromile, L.A.; Shapiro, L.H. Molecular mechanisms regulating CD13-mediated adhesion. Immunology 2014, 142, 636–647.
- Licona-Limón, I.; Garay-Canales, C.A.; Muñoz-Paleta, O.; Ortega, E. CD13 mediates phagocytosis in human monocytic cells. J. Leukoc. Biol. 2015, 98, 85–98.
- Mina-Osorio, P.; Winnicka, B.; O’Conor, C.; Grant, C.L.; Vogel, L.K.; Rodriguez-Pinto, D.; Holmes, K.V.; Ortega, E.; Shapiro, L.H. CD13 is a novel mediator of monocytic/endothelial cell adhesion. J. Leukoc. Biol. 2008, 84, 448–459.
- Mina-Osorio, P.; Shapiro, L.H.; Ortega, E. CD13 in cell adhesion: Aminopeptidase N (CD13) mediates homotypic aggregation of monocytic cells. J. Leukoc. Biol. 2006, 79, 719–730.
- Pérez-Figueroa, E.; Álvarez-Carrasco, P.; Ortega, E. Crosslinking of membrane CD13 in human neutrophils mediates phagocytosis and production of reactive oxygen species, neutrophil extracellular traps and proinflammatory cytokines. Front. Immunol. 2022, 13, 6681.
- Mendoza-Coronel, E.; Ortega, E. Macrophage Polarization Modulates FcγR- and CD13-Mediated Phagocytosis and Reactive Oxygen Species Production, Independently of Receptor Membrane Expression. Front. Immunol. 2017, 8, 303.
- Cheng, H.C.; Abdel-Ghany, M.; Pauli, B.U. A Novel Consensus Motif in Fibronectin Mediates Dipeptidyl Peptidase IV Adhesion and Metastasis. J. Biol. Chem. 2003, 278, 24600–24607.
- Klemann, C.; Wagner, L.; Stephan, M.; von Hörsten, S. Cut to the chase: A review of CD26/dipeptidyl peptidase-4’s (DPP4) entanglement in the immune system. Clin. Exp. Immunol. 2016, 185, 1–21.
- Iwata, B.S.; Morimoto, C. CD26/Dipeptidyl Peptidase IV in Context. J. Exp. Med. 1999, 190, 301–305.
- Engel, M.; Hoffmann, T.; Wagner, L.; Wermann, M.; Heiser, U.; Kiefersauer, R.; Huber, R.; Bode, W.; Demuth, H.-U.; Brandstetter, H. The crystal structure of dipeptidyl peptidase IV (CD26) reveals its functional regulation and enzymatic mechanism. Proc. Natl. Acad. Sci. USA 2003, 100, 5063–5068.
- Nistala, R.; Savin, V. Diabetes, hypertension, and chronic kidney disease progression: Role of DPP4. Am. J. Physiol. Physiol. 2017, 312, F661–F670.
- Raha, A.A.; Chakraborty, S.; Henderson, J.; Mukaetova-Ladinska, E.; Zaman, S.; Trowsdale, J.; Raha-Chowdhury, R. Investigation of CD26, a potential SARS-CoV-2 receptor, as a biomarker of age and pathology. Biosci. Rep. 2020, 40, 20203092.
- Abbott, C.A.; McCaughan, G.W.; Baker, E.; Sutherland, G.R. Genomic organization, exact localization, and tissue expression of the human CD26 (dipeptidyl peptidase IV) gene. Immunogenetics 1994, 40, 331–338.
- Salgado, F.J.; Vela, E.; Martın, M.; Franco, R.; Nogueira, M.; Cordero, O.J. Mechanisms of CD26/dipeptidyl peptidase IV cytokine-dependent regulation on human activated lymphocytes. Cytokine 2000, 12, 1136–1141.
- Fan, H.; Tansi, F.L.; Weihofen, W.A.; Böttcher, C.; Hu, J.; Martinez, J.; Saenger, W.; Reutter, W. Molecular mechanism and structural basis of interactions of dipeptidyl peptidase IV with adenosine deaminase and human immunodeficiency virus type-1 transcription transactivator. Eur. J. Cell Biol. 2012, 91, 265–273.
- Morimoto, C.; Schlossman, S.F. The structure and function of CD26 in the T-cell immune response. Immunol. Rev. 1998, 161, 55–70.
- Kameoka, J.; Tanaka, T.; Nojima, Y.; Schlossman, S.F.; Morimoto, C. Direct Association of Adenosine Deaminase with a T Cell Activation Antigen, CD26. Science 1993, 261, 466–469.
- Lambeir, A.-M.; Durinx, C.; Scharpé, S.; De Meester, I. Dipeptidyl-Peptidase IV from Bench to Bedside: An Update on Structural Properties, Functions, and Clinical Aspects of the Enzyme DPP IV. Crit. Rev. Clin. Lab. Sci. 2003, 40, 209–294.
- A Havre, P.; Dang, L.H.; Ohnuma, K.; Iwata, S.; Morimoto, C.; Dang, N.H. CD26 Expression on T-Anaplastic Large Cell Lymphoma (ALCL) Line Karpas 299 is associated with increased expression of Versican and MT1-MMP and enhanced adhesion. BMC Cancer 2013, 13, 517.
- Gine, S.; Mariño, M.; Mallol, J.; Canela, E.I.; Morimoto, C.; Callebaut, C.; Hovanessian, A.; Casadó, V.; Lluis, C.; Franco, R. Regulation of epithelial and lymphocyte cell adhesion by adenosine deaminase-CD26 interaction. Biochem. J. 2002, 361, 203–209.
- Samavati, L.; Uhal, B.D. ACE2, Much More Than Just a Receptor for SARS-COV-2. Front. Cell. Infect. Microbiol. 2020, 10, 317.
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.-C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ. Res. 2020, 126, 1456–1474.
- Zhang, Q.; Gefter, J.; Sneddon, W.B.; Mamonova, T.; Friedman, P.A. ACE2 interaction with cytoplasmic PDZ protein enhances SARS-CoV-2 invasion. iScience 2021, 24, 102770.
- Kliche, J.; Kuss, H.; Ali, M.; Ivarsson, Y. Cytoplasmic short linear motifs in ACE2 and integrin β3 link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal 2021, 14, 1117.
- Hrenak, J.; Simko, F. Renin–angiotensin system: An important player in the pathogenesis of acute respiratory distress syndrome. Int. J. Mol. Sci. 2020, 21, 8038.
- Bourgonje, A.R.; Abdulle, A.E.; Timens, W.; Hillebrands, J.L.; Navis, G.J.; Gordijn, S.J.; Bolling, M.C.; Dijkstra, G.; Voors, A.A.; Osterhaus, A.D.; et al. Angiotensin-converting enzyme 2 (ACE2), SARS-CoV-2 and the pathophysiology of coronavirus disease 2019 (COVID-19). J. Pathol. 2020, 251, 228–248.
- Brandi, M.L. Are sex hormones promising candidates to explain sex disparities in the COVID-19 pandemic? Rev. Endocr. Metab. Disord. 2022, 23, 171–183.
- Purkayastha, A.; Sen, C.; Garcia Jr, G.; Langerman, J.; Shia, D.W.; Meneses, L.K.; Vijayaraj, P.; Durra, A.; Koloff, C.R.; Freund, D.R.; et al. Direct Exposure to SARS-CoV-2 and Cigarette Smoke Increases Infection Severity and Alters the Stem Cell-Derived Airway Repair Response. Cell Stem Cell 2020, 27, 869–875.e4.
- Simõese Silva, A.C.; Silveira, K.D.; Ferreira, A.J.; Teixeira, M.M. ACE2, angiotensin-(1-7) and Mas receptor axis in inflammation and fibrosis. Br. J. Pharmacol. 2013, 169, 477–492.
More
Information
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
3 times
(View History)
Update Date:
03 Apr 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




