Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Guangqiang Zhang | -- | 2617 | 2023-03-02 08:37:32 | | | |
| 2 | Rita Xu | Meta information modification | 2617 | 2023-03-02 08:42:29 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Kou, H.; Zhang, Z.; Yang, Y.; Wei, C.; Xu, L.; Zhang, G. Aegilops tauschii and the Utilization in Wheat. Encyclopedia. Available online: https://encyclopedia.pub/entry/41806 (accessed on 16 January 2026).
Kou H, Zhang Z, Yang Y, Wei C, Xu L, Zhang G. Aegilops tauschii and the Utilization in Wheat. Encyclopedia. Available at: https://encyclopedia.pub/entry/41806. Accessed January 16, 2026.
Kou, Hongyun, Zhenbo Zhang, Yu Yang, Changfeng Wei, Lili Xu, Guangqiang Zhang. "Aegilops tauschii and the Utilization in Wheat" Encyclopedia, https://encyclopedia.pub/entry/41806 (accessed January 16, 2026).
Kou, H., Zhang, Z., Yang, Y., Wei, C., Xu, L., & Zhang, G. (2023, March 02). Aegilops tauschii and the Utilization in Wheat. In Encyclopedia. https://encyclopedia.pub/entry/41806
Kou, Hongyun, et al. "Aegilops tauschii and the Utilization in Wheat." Encyclopedia. Web. 02 March, 2023.
Copy Citation
Aegilops tauschii is one of the malignant weeds that affect wheat production and is also the wild species ancestor of the D genome of hexaploid wheat (Triticum aestivum, AABBDD). It contains many disease resistance genes that have been lost in the long-term evolution of wheat and is an important genetic resource for the mining and utilization of wheat disease resistance genes.
wheat
Aegilops tauschii
genetics
genomics
1. Introduction
As one of the major crops, wheat is the main grain for about one third of the world’s population, and its yield is of great significance for alleviating global hunger [1]. According to the FAO, pests and diseases cause 20–40% of global food crop losses and losses of USD 220 billion in agricultural trade every year [2]. In the long-term natural selection and artificial selection, wheat has lost many excellent disease resistance genes [3][4], resulting in a single genetic background of wheat.
Aegilops tauschii belongs to the genus Aegilops in the Triticeae family of Poaceae [5]. It is an annual weed in wheat fields and is also a relative plant of wheat and the donor species of the D genome during the evolution of common hexaploid wheat [6][7]. Compared with the D genome of wheat, Aegilops tauschii has a richer genetic diversity and contains many stress resistance, disease resistance, and insect resistance genes, among many other excellent genes, which are an important breeding resource for wheat breeders to improve wheat traits as well as disease resistance and stress resistance [8][9][10][11]. Therefore, fully exploring and utilizing the excellent disease resistance genes in Aegilops tauschii are of great value for wheat disease resistance breeding.
2. The Relationship between Aegilops tauschii and Wheat
Aegilops tauschii (DD, 2n = 14), which originates from West Asia, is mainly distributed in the Middle East, Europe, West Asia, and other places [12][13]. In China, Aegilops tauschii is mainly distributed in Xinjiang and the Yellow River Basin (including Shaanxi and Henan Provinces) [14][15]. There are two main lines of introducing Aegilops tauschii into the Yellow River Basin of China: first, Middle East→Russia→Xinjiang, China→Yellow River Basin into China; second, it was directly imported from the Middle East via the ancient Silk Road without passing through Xinjiang [16] (Figure 1a,b). As the donor of the D genome of wheat, Aegilops tauschii has a wider distribution area and higher genetic diversity than common hexaploid wheat. The addition of the D genome makes hexaploid wheat more adaptable to continental climates, which has laid a solid foundation for the large-scale cultivation of wheat [17].
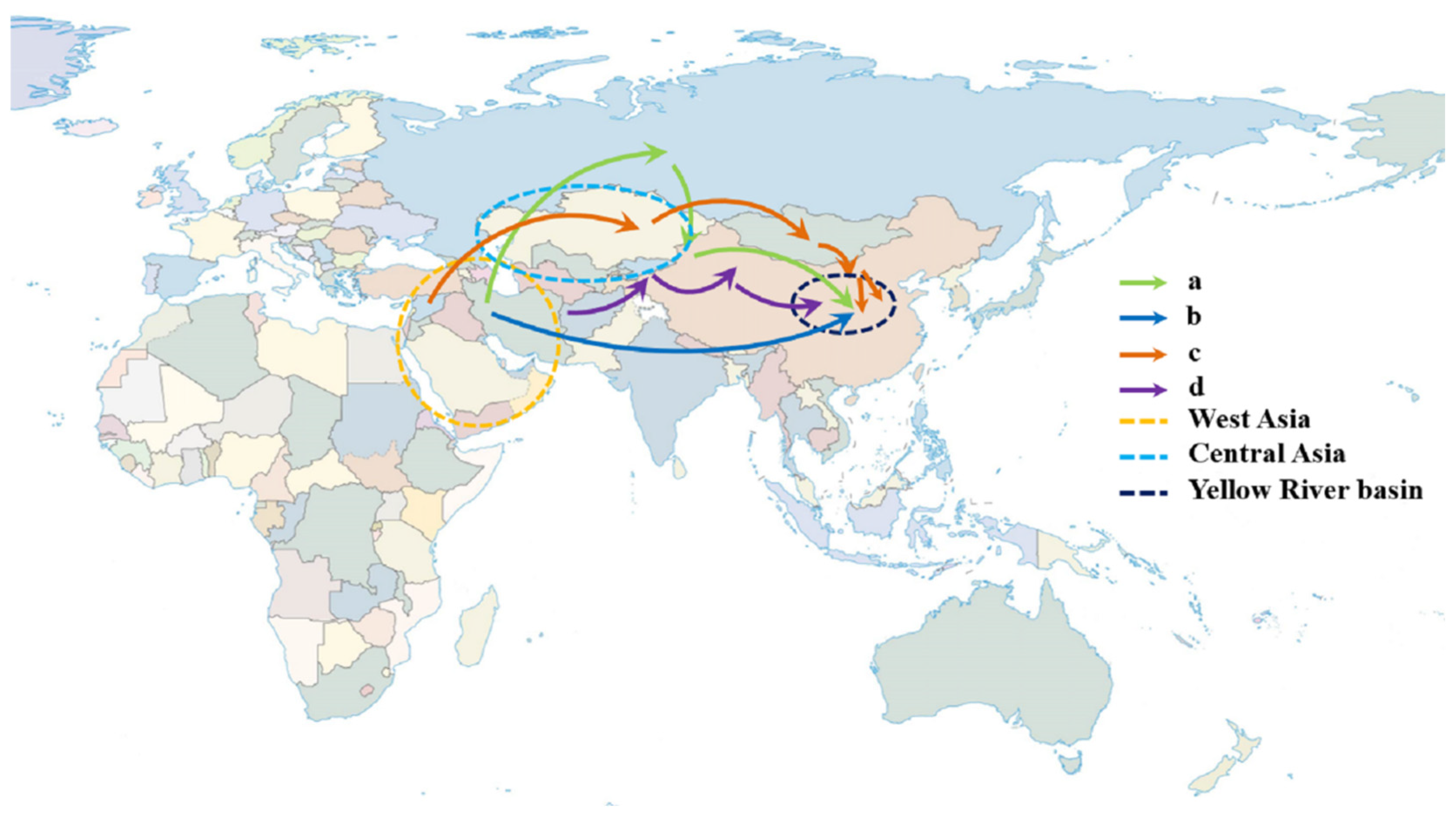
Figure 1. Road map of the introduction of Aegilops tauschii and wheat into China. The route of Aegilops tauschii introduced into China: (a) Middle East→Russia→Xinjiang, China→Yellow River Basin; (b) the ancient Silk Road in the Middle East; (c) northwest of West Asia→Eurasia→southern Siberia and Mongolia→Middle and Lower Yellow River Region; (d) Afghanistan or Central Asia Oasis →Northern Xinjiang→Yellow River Basin.
Wheat originated in the fertile crescent of the Middle East. The first domesticated wild wheat was Triticum monococcum, followed by cultivated emmer wheat (Triticum turgidum); finally, the common wheat (Triticum aestivum) was formed through natural hybridization between cultivated emmer wheat and Aegilops tauschii along the Caspian Sea coast [18]. As for how wheat evolved from diploid wheat to the present allohexaploid wheat, there are two theories: one is formation by direct homoploid hybridization. Marcussen et al. found that approximately 6.5 million years ago, the wheat lineage (Triticum and Aegilops) began to differentiate from a common ancestor into the A and B genome lineages. About 5.5 million years ago, the first hybridization occurred between the A and B genome lineages and led to the origin of the D genome lineage. Furthermore, the second hybridization between the A genome donor Triticum urartu (AA) and a related species (BB) of Aegilops speltodies occurred approximately 0.8 million years ago, resulting in allotetraploid emmer wheat (Triticum turgidum; AABB), which then acclimated to cultivated tetraploid wheat, being crossed again with the D genome donor Aegilops tauschii (DD) about 0.4 million years ago to form hexaploid wheat; finally, it was acclimated to Triticum aestivum (AABBDD) [19]. In addition, Li et al. re-evaluated the homoploid hybrid origin of Aegilops tauschii. Based on the whole chloroplast genome sequence, they analyzed the neighbor joining tree of the Triticum–Aegilops complex and found that the chloroplast topology reveals that Aegilops tauschii is cladistically nested between the A and remaining S * and M genomes. They gave two possible explanations, i.e., the chloroplast capture model and the ancestry capture model. Therefore, they clearly pointed to a more complex history of Aegilops tauschii than that proposed by Marcussen et al. [19], one that may have involved multiple rounds of both recent and ancient hybridizations [20]. Then, Aegilops tauschii hybridized with tetraploid wheat to form hexaploid wheat. According to Alison et al., wheat was introduced into China in at least two ways. The first is that wheat came from the northwest of West Asia, moving through Eurasia, southern Siberia, and Mongolia to the middle and lower reaches of the Yellow River; the second is that wheat came from West Asia, potentially moving through the Afghanistan or Central Asian oases and northern Xinjiang into China’s Yellow River Basin, rather than the Eurasian steppe [21] (Figure 1c,d). China is the secondary origin center of wheat, mainly including three wheat subspecies: Yunnan wheat (Triticum aestivum ssp. yunnanense King), Tibet semi-wild wheat (Triticum aestivum ssp. tibetanum Shao), and Xinjiang wheat (Triticum aestivum ssp. Petropavlovsk yi) [22].
Since the domestication of wheat, yield improvement has always been the focus of wheat research. By improving cultivation measures and breeding techniques, the yield per unit area of wheat has been greatly increased [23][24][25][26][27][28][29][30][31][32]. Among them, enhancing wheat disease resistance has always been an important part of improving wheat yield. Aegilops tauschii contains abundant beneficial genes for disease resistance, insect resistance, cold resistance, and high quality, which is of great significance for improving wheat yield.
By analyzing the relationship between the two species, it has been proved that Aegilops tauschii is an important germplasm resource of wheat. During the long-term evolution process of wheat, the genetic diversity of the D genome has gradually narrowed, resulting in few available genetic resources. However, as the donor of the D genome of wheat, Aegilops tauschii carries abundant excellent genes, and the genetic material between them can be exchanged and recombined, which provides a valuable genetic resource pool for wheat breeding. Since both of these wheat species originated in West Asia, researchers can further enrich wheat germplasm resources by exploring wild wheat and Aegilops tauschii from West Asia.
3. Application of Disease-Resistant Genes from Aegilops tauschii in Wheat Breeding
The utilization of disease resistance genes in Aegilops tauschii is of great significance for expanding wheat disease resistance. Synthetic hexaploid wheat (SHW) is an artificially created hexaploid wheat that can simultaneously introduce genetic variations from tetraploid wheat and Aegilops tauschii, and it has been widely used to expand the genetic diversity of common wheat [33]. The method mainly includes two main steps: First, a hybrid F1 with an ABD genome is produced by direct hybridization of tetraploid wheat with Aegilops tauschii, and then a synthetic hexaploid wheat with an AABBDD genome is obtained through chromosome doubling [33]. Second, the genetic variation in Aegilops tauschii and tetraploid wheat is introduced into common wheat varieties by using the artificial synthetic hexaploid wheat as a bridge and common wheat as a backcross or topcross [34].
3.1. Application of Rust Resistance Genes from Aegilops tauschii in Wheat Breeding
3.1.1. Application of Stripe Rust Resistance Genes from Aegilops tauschii in Wheat Breeding
Yr28 is the first stripe rust resistance gene cloned from Aegilops tauschii. Through map-based cloning results, previous researchers designed resistance co-segregation molecular markers, conducted auxiliary selection, and bred a new variety, Shumai 1675 [17]. The main cultivation processes were as follows: (1) introducing disease resistance genes into synthetic hexaploid wheat; (2) establishing a breeding population; (3) F2 small group mixed selection; (4) F3 small population for molecular marker selection to prevent target gene loss; (5) F5 line focused on the selection of yield-related traits, and molecular marker selection of disease resistance genes [35]. After the above processes, the yield of F5 and its selected line Shumai 1675 increased significantly and showed stripe rust resistance.
3.1.2. Application of Leaf Rust Resistance Genes from Aegilops tauschii in Wheat Breeding
Lr21 is the first powdery mildew resistance gene found and successfully cloned from Aegilops tauschii. Thus far, the application of the Lr21 gene in wheat breeding is low. Mebrate et al. [36] used 31 Pt races to detect 36 wheat cultivars from Ethiopia and Germany and found that Sirbo and Granny contained Lr21. Gebrewahid et al. [37] identified 83 wheat varieties and 36 lines with known leaf rust resistance (Lr) genes from three provinces in China. There were 41 cultivars containing leaf rust resistance (Lr) genes, but only Wanmai 47 contained Lr21. Khakimova et al. [38] studied 36 synthetic hexaploid wheat varieties from Russia and identified 11 materials containing Lr21. Zhang et al. [39] identified and analyzed 46 Chinese landraces and found that only Baiheshang contained Lr21.
Lr22a has broad-spectrum resistance to wheat leaf rust, but it has not been widely used in production due to differences in varieties from different regions. Khakimova et al. [38] studied 36 synthetic hexaploid wheat varieties from Russia and found that three of them contained Lr22a. Huang et al. identified and analyzed 36 wheat production varieties in Gansu Province and found that the varieties Huining 15, Lantian 37, and Longjian 113 showed resistance to all Lr22a non-toxic races, indicating that these three materials may contain Lr22a [40]. However, Atia et al. [41] identified and analyzed 50 wheat varieties in Egypt and successfully identified 21 Lr genes, and all wheat varieties contained Lr22a.
Although Lr32 has not been cloned successfully, it was found that the disease-resistant wheat varieties contained this gene in actual production identification. Zhao et al. identified 23 Chinese wheat microcore collections, and the five core germplasms of Tongjiaba wheat, Honghua wheat, Kefeng 3, Atlas66, and Golden wheat contained the Lr32 gene [42]. Hanaa et al. [43] identified leaf rust resistance in 10 Egyptian spring wheat varieties at the seedling stage and found that Sids12 and Sakha93 contained Lr32. Bahar et al. [44] used SSR markers of 13 resistance genes to identify 57 wheat lines and found that all 57 lines contained Lr32. Atia et al. [41] identified and analyzed 50 wheat varieties in Egypt and successfully identified 21 Lr genes, and all wheat varieties contained Lr32.
Lr39 is a powdery mildew resistance gene of wheat at the seedling stage, and it has certain development value [45]. Hanaa et al. [43] identified leaf rust resistance in 10 Egyptian spring wheat varieties at the seedling stage and found that Miser1 and Miser2 contained Lr39. Atia et al. [41] identified and analyzed 50 wheat varieties in Egypt and successfully identified 21 Lr genes, and 42 wheat varieties contained Lr39. Wang et al. identified the leaf rust resistance of 71 important wheat production varieties in Henan Province and found that four cultivars contained Lr39 [46].
As early as 1991, Lr42 was transferred from Aegilops tauschii to common wheat through hybridization by the Wheat Germplasm Resources Center (WGRC) of Kansas State University in the United States, and the KS91WGRC11 wheat line was developed [47]. Subsequently, the International Maize and Wheat Improvement Center (CIMMYT) widely applied the disease resistance genes in this line to breeding materials. Through the identification and analysis of 103 wheat varieties (lines) of CIMMYT and 35 control varieties containing known leaf rust resistance genes, Han et al. found that 11 CIMMYT wheat varieties may contain Lr42 [48]. Liu et al. tested 66 wheat varieties approved by Qinghai Province and found that 23 varieties contained Lr42, accounting for 34.85% [49]. Among 52,943 CIMMYT lines or varieties sequenced by GBS, 5121 pedigrees contained Lr42 [50].
3.1.3. Application of Stem Rust Resistance Genes from Aegilops tauschii in Wheat Breeding
Sr33 is an important gene for resistance to the physiological race Ug99 of stem rust, and its wide application is of great significance to reduce the harm of stem rust. Ma et al. conducted SSR detection on 58 spring wheat varieties resistant to Ug99 introduced at home and abroad and 18 main wheat varieties in Heilongjiang Province and found that only one spring wheat variety material resistant to Ug99 and three main wheat varieties in Heilongjiang Province contained the Sr33 gene [51].
Periyannan et al. [34] found that Sr45 was effective against Puccinia graminis f. sp. tritici races prevalent in small populations in Australia and South Africa and the Ug99 race group, but the related detection was lower.
Kokhmetova and Atishova found that only the Sr46 gene existed in the 338-K1-1//ANB/BUC/3/GS50A/4/422/5/BAYRAKTAR line when detecting 88 cultivars of spring soft wheat in Kazakhstan [52].
3.2. Application of Powdery Mildew Resistance Genes from Aegilops tauschii in Wheat Breeding
Pm2a, as a successfully cloned gene from Aegilops tauschii, is of great significance in wheat breeding for powdery mildew resistance. Švec et al. [53] identified the Pm2 gene in 32 Polish wheat varieties. Agnieszka et al. [54] identified seven wheat varieties from Europe by establishing a multiplex PCR reaction and found that all wheat varieties contained the Pm2 gene. Jimai 22 has been approved and widely promoted in Shandong Province and the northern part of Huanghuai; as of the summer harvest in 2020, the cumulative promotion area was 20 million hm2. Liangxing 66 has been promoted and planted in Shandong, central and southern Hebei, southern Shanxi, and Anyang, Henan, to the north of the Huanghuai winter wheat region [55]. Through genetic analysis and molecular marker detection, the above two varieties were found to carry the wheat powdery mildew resistance gene Pm2 [56]. With the increase in the utilization frequency of Pm2 in production, the frequency of the corresponding virulent species variation is also rising, resulting in an increasing risk of overcoming Pm2 resistance.
With the acceleration of variety replacement, the varieties containing Pm19 in actual production have gradually increased. Li et al. [57] identified 23 white powdery mildew-resistant materials and found that only one material contained Pm19. According to the identification results, Pm19 was considered to have low resistance and should be used in combination with other resistance genes. Shi et al. [58] identified 61 reserve varieties of powdery mildew in China and found that 21 wheat varieties contained powdery mildew resistance genes, and four of them contained Pm19.
Although Pm34 has not been cloned, it has been identified to contain this gene in wheat in actual production. Li et al. [59] identified 42 Yunnan wheat varieties using 20 wheat powdery mildew strains with different toxicity profiles and found that four varieties contained Pm34. Wang et al. [60] analyzed 305 wheat germplasm resources at home and abroad and found that 95 wheat varieties contained Pm34, accounting for 31.15%, including Lumai 5, Yanzhan 4110, Fengsheng 3, Jimai 22, CA9719, and Azulon.
El-Shamy et al. [61] used 12 Egyptian wheat varieties to identify the virulence of 52 powdery mildew strains and found that wheat varieties containing the Pm35 gene had higher disease resistance. However, in actual production, there are few wheat varieties containing Pm35. Through toxicity monitoring and annual dynamic change analysis of wheat powdery mildew populations in Shaanxi Province, China, Liu et al. found that NCD3 wheat varieties containing Pm35 had higher disease resistance [62]. Yan et al. identified 371 wheat materials from Hebei Province and found that only Pubing 01 contained the Pm35 gene [63].
Due to the late discovery and cloning of Pm58, only the germplasm lines U6714-A-011 (Reg.No.GP-1023, PI682090) and U6714-B-056 (Reg.No.GP-1022, PI 682089) of the new powdery mildew resistance gene Pm58 cultivated by Michigan State University using TA1662 and KS05HW14 are currently available, but the wheat yields of these two germplasm lines need to be improved [64].
3.3. Application of Other Disease-Resistant Genes from Aegilops tauschii in Wheat Breeding
Because the genetic research on wheat septoria tritici blotch resistance genes are relatively slow, Stb5, as the only localized gene of wheat septoria tritici blotch in Aegilops tauschii, has not been widely studied and utilized in production [65].
Tsr3 is one of the four genes for wheat brown spot disease resistance officially mapped in Aegilops tauschii [66]. The research on Tsr3 is less than that on wheat resistance to wheat septoria tritici blotch, and relevant production research reports have not been found yet.
References
- Appels, R.; Eversole, K.; Feuillet, C.; Keller, B.; Rogers, J.; Stein, N.; Pozniak, C.J.; Choulet, F.; Distelfeld, A.; Poland, J.; et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191.
- The Food and Agriculture Organization of the United Nations Home Page. Available online: https://www.fao.org/director-general/news/news-article/zh/c/1301879/ (accessed on 15 November 2022).
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.L.; Freeling, M.; Wang, X.W. Gene retention, fractionation and subgenome differences in polyploid plants. Nat. Plants 2018, 4, 258–268.
- Liang, Y.M.; Liu, H.J.; Yan, J.B.; Tian, F. Natural variation in crops: Realized understanding, continuing promise. Annu. Rev. Plant Biol. 2021, 72, 357–385.
- Dvorak, J.; Luo, M.C.; Yang, Z.L.; Zhang, H.B. The structure of the Aegilops tauschii genepool and the evolution of hexaploid wheat. Theor. Appl. Genet. 1998, 97, 657–670.
- Wang, J.R.; Luo, M.C.; Chen, Z.X.; You, F.M.; Wei, Y.M.; Zheng, Y.L.; Dvorak, J. Aegilops tauschii single nucleotide polymorphisms shed light on the origins of wheat D-genome genetic diversity and pinpoint the geographic origin of hexaploid wheat. New Phytol. 2013, 198, 925–937.
- Huang, Z.F.; Sui, B.F.; Zhang, C.X.; Huang, H.J.; Wei, S.H. The basis of resistance mechanism to mesosulfuron-methyl in Tausch’s goatgrass (Aegilops tauschii Coss.). Pestic. Biochem. Physiol. 2019, 155, 126–131.
- Hegde, S.G.; Valkoun, J.; Waines, J.G. Genetic diversity in wild and weedy Aegilops, Amblyopyrum, and Secale species—A preliminary survey. Crop Sci. 2002, 42, 608–614.
- Zhang, D.L.; Zhou, Y.; Zhao, X.P.; Lü, L.L.; Zhang, C.C.; Li, J.H.; Sun, G.L.; Li, S.P.; Song, C.P. Development and utilization of introgression lines using synthetic octaploid wheat (Aegilops tauschii × Hexaploid wheat) as donor. Front. Plant Sci. 2018, 9, 1113.
- Masahiro, K. An update of recent use of Aegilops species in wheat Breeding. Front. Plant. Sci. 2019, 10, 585.
- Zhou, Y.; Bai, S.L.; Li, H.; Sun, G.L.; Zhang, D.L.; Ma, F.F.; Zhao, X.P.; Nie, F.; Li, J.Y.; Chen, L.Y.; et al. Introgressing the Aegilops tauschii genome into wheat as a basis for cereal improvement. Nat. Plants 2021, 7, 774–786.
- Wei, H.T.; Li, J.; Peng, Z.S.; Lu, B.R.; Zhao, Z.J.; Yang, W.Y. Relationships of Aegilops tauschii revealed by DNA fingerprits: The evidence for agriculture exchange between China and the West. Prog. Nat. Sci.-Mater. 2008, 18, 1525–1531.
- Maciej, M.; Michał, T.K.; Joanna, M.; Halina, W. Aegilops tauschii accessions with geographically diverse origin show differences in chromosome organization and polymorphism of molecular markers linked to leaf rust and powdery mildew resistance genes. Front. Plant Sci. 2017, 8, 1149.
- Ward, R.W.; Yang, Z.L.; Kim, H.S.; Yen, C. Comparative analyses of RFLP diversity in landraces of Triticum aestivum and collections of T. tauschii from China and Southwest Asia. Theor. Appl. Genet. 1998, 96, 312–318.
- Su, Q.; Liu, L.X.; Zhao, M.Y.; Zhang, C.C.; Zhang, D.L.; Li, Y.Y.; Li, S.P. The complete chloroplast genomes of seventeen Aegilops tauschii: Genome comparative analysis and phylogenetic inference. PeerJ 2020, 8, e8678.
- Zhao, X.P.; Zhou, Y.; Lü, L.L.; Li, S.P.; Zhang, D.L. Genetic diversity of Aegilops tauschii Coss. and its utilization in improving common wheat. Biol. Bull. 2019, 35, 181–189. (In Chinese)
- Zhang, C.Z.; Huang, L.; Zhang, H.F.; Hao, Q.Q.; Lyu, B.; Wang, M.N.; Epstein, L.; Liu, M.; Kou, C.L.; Qi, J.; et al. An ancestral NB-LRR with duplicated 3′UTRs confers stripe rust resistance in wheat and barley. Nat. Commun. 2019, 10, 4023.
- Rahman, S.; Islam, S.; Yu, Z.T.; She, M.Y.; Nevo, E.; Ma, W.J. Current progress in understanding and recovering the wheat genes lost in evolution and domestication. Int. J. Mol. Sci. 2020, 21, 5836.
- Marcussen, T.; Sandve, S.R.; Heier, L.; Spannagl, M.; Pfeifer, M.; The International Wheat Genome Sequencing Consortium; Jakobsen, K.S.; Wulff, B.B.H.; Steuernagel, B.; Mayer, K.F.X.; et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science 2014, 345, 1250092.
- Li, L.F.; Liu, B.; Olsen, K.M.; Wendel, J.F. A re-evaluation of the homoploid hybrid origin of Aegilops tauschii, the donor of the wheat D-subgenome. New Phytol. 2015, 208, 4–8.
- Alison, B.; Peter, W.J.; John, D. The origins of wheat in China and potential pathways for its introduction: A review. Quat. Int. 2014, 348, 158–168.
- Liu, J.; Yao, Y.Y.; Xin, M.M.; Peng, H.R.; Ni, Z.F.; Sun, Q.X. Shaping polyploid wheat for success: Origins, domestication, and the genetic improvement of agronomic traits. J. Integr. Plant Biol. 2021, 64, 536–563.
- Ben, T.; Megan, N.H.; Elizabeth, S.D.; Peacock, W.J. The molecular basis of vernalization-induced flowering in cereals. Trends Plant Sci. 2007, 12, 352–357.
- Wang, Y.H.; Liu, H.; Huang, Y.; Wang, J.F.; Wang, Z.Z.; Gu, F.X.; Xin, M.H.; Kang, G.Z.; Feng, W.; Guo, T.C. Effects of cultivation management on the winter wheat grain yield and water utilization efficiency. Sci. Rep. 2019, 9, 12733.
- Hao, Z.M.; Geng, M.M.; Hao, Y.R.; Zhang, Y.; Zhang, L.J.; Wen, S.M.; Wang, R.H.; Liu, G.R. Screening for differential expression of genes for resistance to Sitodiplosis mosellana in bread wheat via BSR-seq analysis. Theor. Appl. Genet. 2019, 132, 3201–3221.
- Li, J.B.; Dundas, I.; Dong, C.M.; Li, G.R.; Trethowan, R.; Yang, Z.J.; Hoxha, S.; Zhang, P. Identification and characterization of a new stripe rust resistance gene Yr83 on rye chromosome 6R in wheat. Theor. Appl. Genet. 2020, 133, 1095–1107.
- Wang, H.W.; Sun, S.L.; Ge, W.Y.; Zhao, L.F.; Hou, B.Q.; Wang, K.; Lyu, Z.F.; Chen, L.Y.; Xu, S.S.; Guo, J.; et al. Horizontal gene transfer of Fhb7 from fungus underlies Fusarium head blight resistance in wheat. Science 2020, 368, eaba5435.
- Banach, J.K.; Majewska, K.; ŻukGołaszewska, K. Effect of cultivation system on quality changes in durum wheat grain and flour produced in North-Eastern Europe. PLoS ONE 2021, 16, e0236617.
- Wang, H.M.; Zheng, J.; Fan, J.L.; Zhang, F.C.; Huang, C.H. Grain yield and greenhouse gas emissions from maize and wheat fields under plastic film and straw mulching: A meta-analysis. Field Crop. Res. 2021, 270, 108210.
- Yang, H.K.; Xiao, Y.; Zhang, X.; Huang, X.L.; Fan, G.Q. Maize straw mulching with uniconazole application increases the tillering capacity and grain yield of dryland winter wheat (Triticum aestivum L.). Field Crop. Res. 2022, 284, 108573.
- Yang, X.L.; Wang, G.Y.; Chen, Y.Q.; Sui, P.; Pacenka, S.; Steenhuis, T.S.; Siddique, K.H.M. Reduced groundwater use and increased grain production by optimized irrigation scheduling in winter wheat–summer maize double cropping system—A 16-year field study in North China Plain. Field Crop. Res. 2022, 275, 108364.
- Zhang, X.Y.; Jia, H.Y.; Li, T.; Wu, J.Z.; Nagarajan, R.; Lei, L.; Powers, C.; Kan, C.C.; Hua, W.; Liu, Z.Y.; et al. TaCol-B5 modifies spike architecture and enhances grain yield in wheat. Science 2022, 376, 180–183.
- Li, A.L.; Liu, D.C.; Yang, W.Y.; Kishii, M.; Mao, L. Synthetic hexaploid wheat: Yesterday, today, and tomorrow. Engineering 2018, 4, 552–558.
- Periyannan, S.; Bansal, U.; Bariana, H.; Deal, K.; Luo, M.C.; Dvorak, J.; Lagudah, E. Identification of a robust molecular marker for the detection of the stem rust resistance gene Sr45 in common wheat. Theor. Appl. Genet. 2014, 127, 947–955.
- Hao, M.; Zhang, L.Q.; Huang, L.; Ning, S.Z.; Yuan, Z.W.; Jiang, B.; Yan, Z.H.; Wu, B.H.; Zheng, Y.L.; Liu, D.C. Genetic improvement of synthesized hexaploid wheat in breeding. J. Plant Genet. Resour. 2022, 23, 40–48. (In Chinese)
- Mebrate, S.A.; Dehne, H.W.; Pillen, K.; Oerke, E.C. Postulation of seedling leaf rust resistance genes in selected Ethiopian and German bread wheat cultivars. Crop. Sci. 2008, 48, 507–516.
- Gebrewahid, T.W.; Yao, Z.J.; Yan, X.C.; Gao, P.; Li, Z.F. Identification of leaf rust resistance genes in Chinese common wheat cultivars. Plant Dis. 2017, 101, 1729–1737.
- Khakimova, A.G.; Gultyaeva, E.I.; Mitrofanova, O.P. Resistance of synthetic hexaploid wheat to the leaf rust pathogen. Proc. Appl. Bot. Genet. Breed. 2018, 179, 125–136.
- Zhang, P.P.; Gebrewahid, W.T.; Zhou, Y.; Li, Q.L.; Li, Z.F.; Liu, D.Q. Seedling and adult plant resistance to leaf rust in 46 chinese bread wheat landraces and 39 wheat lines with known Lr genes. J. Integr. Agric. 2019, 18, 1014–1023.
- Huang, J.; Jin, S.L.; Cao, S.Q.; Jia, Q.Z.; Luo, H.S.; Zhang, B.; Sun, Z.Y.; Wang, X.M. Postulation of leaf rust resistance genes of 36 wheat cultivars developed in Gansu and their resistance evaluation at adult plant stage. Plant Prot. 2020, 46, 171–177, 188. (In Chinese)
- Atia, M.A.M.; El-Khateeb, E.A.; Abd, E.-M.R.M.; Abou-Zeid, M.A.; Salah, A.; Abdel-Hamid, A.M.E. Mining of leaf rust resistance genes content in Egyptian bread wheat collection. Plants 2021, 10, 1378.
- Zhao, L.N.; Ren, X.D.; Hu, Y.Y.; Zhang, T.; Zhang, N.; Yang, W.X.; Liu, D.Q. Evaluation of wheat leaf rust resistance of 23 Chinese wheat mini-core collections. Sci. Agric. Sin. 2013, 46, 441–450. (In Chinese)
- Hanaa, A.; Eman, E.A.; Walid, E.O. Molecular markers and postulation study of leaf rust resistance genes in various Egyptian wheat cultivars. Biotechnol. J. Int. 2017, 20, 1–13.
- Bahar, A.; Iqbal, M.; Aqib, I.; Mian, A.A.; Iram, M.; Muhammad, H. Molecular charactarization of wheat advanced lines for leaf rust resistant genes using SSR markers. Microb. Pathog. 2018, 123, 348–352.
- Chen, W.Q.; Qin, Q.M. Studies on utilization of worldwide known genes for leaf rust resistance of wheat in China. Sci. Agric. Sin. 2002, 7, 794–801. (In Chinese)
- Wang, W.X.; Zhang, M.Y.; Dong, R.; Zhang, P.P.; Zhang, J.Y.; Li, Z.F.; Liu, D.Q. Identification of leaf rust resistance of 71 wheat cultivars from Henan Province. J. Triticeae Crops 2022, 42, 279–288. (In Chinese)
- Cox, T.S.; Sears, R.G.; Gill, B.S.; Jellen, E.N. Registration of KS91WGRC11, KS92WGRC15, and KS92WGRC23 leaf rust-resistant hard red winter wheat germplasms. Crop. Sci. 1994, 34, 546–547.
- Han, Y.; He, Z.h.; Xia, X.C.; Li, X.; Li, Z.F.; Liu, D.Q. Seedling and slow rusting resistances to leaf rust in CIMMYT wheat lines. Acta Agron. Sin. 2011, 37, 1125–1133. (In Chinese)
- Liu, T.; Wu, L.J.; Gan, X.L.; Zhang, B.; Liu, B.L.; Chen, W.J.; Zhang, L.Q.; Liu, D.C.; Zhang, H.G. Molecular identification of leaf rust resistance genes in bread wheat cultivars released in Qinghai Province. Acta Agric. Boreali-Occident. Sin. 2018, 27, 1112–1118. (In Chinese)
- Lin, G.F.; Chen, H.; Tian, B.; Sehgal, S.K.; Singh, L.; Xie, J.Z.; Rawat, N.; Juliana, P.; Singh, N.; Shrestha, S.; et al. Cloning of the broadly effective wheat leaf rust resistance gene Lr42 transferred from Aegilops tauschii. Nat. Commun. 2022, 13, 3044.
- Ma, Y.; Shao, L.G.; Wang, Y.; Li, C.H.; Che, J.Y.; Gao, F.M.; Zhang, Q.C.; Liu, N.T.; Zhou, D.Y.; Wang, Z.K. Molecular detection of the stem rust resistant gene Sr33 in spring wheat cultivars. J. Triticeae Crops 2013, 33, 34–38. (In Chinese)
- Kokhmetova, A.M.; Atishova, M.N. Identification of sources of resistance to wheat stem using molecular markers. Russ. J. Genet. Appl. Res. 2012, 2, 486–493.
- Švec, M.; Szunics, L.; Slováková, T.; Miklovičová, M.; Tisová, V.; Hauptvogel, P. Identification of genes for resistanceto wheat powdery mildew in Hungarian, Polish and Slovak wheat cultivars. Plant Prot. Sci. 2002, 54, 64–72.
- Agnieszka, T.; Roksana, S.; Dorota, W.; Michał, K.; Jerzy, N.; Przemysław, Ł.K.; Mateusz, P. Identification of powdery mildew Blumeria graminis f. sp. tritici resistance genes in selected wheat varieties and development of Multiplex PCR. Open Chem. 2019, 17, 157–165.
- Jin, Y.L.; Gu, T.; Liu, H.; An, D.G. Research progress on the wheat powdery mildew resistance gene Pm2. Chin. J. Eco-Agric. 2022, 30, 779–786. (In Chinese)
- Huang, J.; ZHAO, Z.H.; Song, F.J.; Wang, X.M.; Xu, H.X.; Huang, Y.; An, D.G.; Li, H.J. Molecular detection of a gene effective against powdery mildew in the wheat cultivar Liangxing 66. Mol. Breed. 2012, 30, 1737–1745.
- Li, J.P.; Jin, S.L.; Cao, S.Q.; Chen, Y.R.; Jin, M.G. Effectivity and evaluation of the resistant genes to wheat powdery mildew in Gansu Province. J. Plant Prot. 2003, 1, 30–34. (In Chinese)
- Shi, W.Q.; Gong, S.J.; Zeng, F.S.; Xiang, L.B.; Yang, L.J. Evaluation of resistance to powdery mildew in 61 Chinese wheat cultivars and postulation of their resistance genes. J. Plant Prot. 2019, 46, 1086–1099. (In Chinese)
- Li, M.J.; Duan, X.Y.; Zhou, Y.L.; Yu, Y.X.; Bi, Y.Q.; Yang, J.H.; Zhang, Q. Postulation of seedlings resistance genes to powdery mildew in wheat commercial cultivars from Yunnan Province. J. Triticeae Crops 2012, 32, 551–556. (In Chinese)
- Wang, X.; Song, P.B.; Wang, X.X.; Yang, M.Y.; Zhou, F.; Lv, D.Y.; Sun, D.J. Identification and evaluation of resistance to stripe rust and powdery mildew of 305 domestic and foreign wheat germplasms. J. Triticeae Crops 2021, 41, 689–698. (In Chinese)
- El-Shamy, M.M.; Emara, H.M.; Mohamed, M.E. Virulence analysis of wheat powdery mildew (Blumeria graminis f. sp. tritici) and effective genes in Middle Delta, Egypt. Plant Dis. 2016, 100, 1927–1930.
- Liu, W.; Wang, Z.H.; Cao, X.R.; Xu, Z.; Shi, Q.Q.; Han, L.P.; Fan, J.R.; Wang, B.T.; Zhou, Y.L. Monitoring and dynamic change of virulence of Blumeria graminis f. sp. tritici population in Shaanxi Province. Plant Prot. 2018, 44, 154–161. (In Chinese)
- Yan, R.; Geng, M.M.; Li, X.J.; An, H.J.; Wen, S.M.; Liu, G.R.; Wang, R.H. Phenotyping and marker-assisted gene identification of powdery mildew resistance in wheat commercial varieties and germplasm resources from Hebei Province. J. Plant Genet. Resour. 2020, 21, 683–705. (In Chinese)
- Wiersma, A.T.; Whetten, R.B.; Zhang, G.R.; Sehgal, S.K.; Kolb, F.L.; Poland, J.A.; Mason, R.E.; Carter, A.H.; Cowger, C.; Olson, E.L. Registration of two wheat germplasm lines fixed for Pm58. J. Plant Regist. 2018, 12, 270–273.
- Lu, J.L.; Fan, Y.H.; Ma, M.S.; Tian, X.J. Progress on discovery and application of wheat septoria tritici blotch resistance genes. J. Shanxi Agric. Sci. 2021, 49, 1393–1399. (In Chinese)
- Liang, S.; Li, X.F. Research progress of wheat leaf blight diseas. Shandong Agric. Sci. 2022, 54, 139–145. (In Chinese)
More
Information
Subjects:
Agronomy
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.2K
Revisions:
2 times
(View History)
Update Date:
02 Mar 2023
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




