Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rodrigo Araújo Lima Rodrigues | -- | 2442 | 2022-12-22 22:00:11 | | | |
| 2 | Conner Chen | Meta information modification | 2442 | 2022-12-26 04:38:27 | | | | |
| 3 | Conner Chen | + 6 word(s) | 2448 | 2022-12-27 02:34:41 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Oliveira, E.G.D.; Carvalho, J.V.R.P.; Botelho, B.B.; Filho, C.A.D.C.; Henriques, L.R.; Azevedo, B.L.D.; Rodrigues, R.A.L. Phycodnaviridae as Giant Viruses of Interest for Biotechnology. Encyclopedia. Available online: https://encyclopedia.pub/entry/39121 (accessed on 08 February 2026).
Oliveira EGD, Carvalho JVRP, Botelho BB, Filho CADC, Henriques LR, Azevedo BLD, et al. Phycodnaviridae as Giant Viruses of Interest for Biotechnology. Encyclopedia. Available at: https://encyclopedia.pub/entry/39121. Accessed February 08, 2026.
Oliveira, Ellen Gonçalves De, João Victor Rodrigues Pessoa Carvalho, Bruna Barbosa Botelho, Clécio Alonso Da Costa Filho, Lethícia Ribeiro Henriques, Bruna Luiza De Azevedo, Rodrigo Araújo Lima Rodrigues. "Phycodnaviridae as Giant Viruses of Interest for Biotechnology" Encyclopedia, https://encyclopedia.pub/entry/39121 (accessed February 08, 2026).
Oliveira, E.G.D., Carvalho, J.V.R.P., Botelho, B.B., Filho, C.A.D.C., Henriques, L.R., Azevedo, B.L.D., & Rodrigues, R.A.L. (2022, December 22). Phycodnaviridae as Giant Viruses of Interest for Biotechnology. In Encyclopedia. https://encyclopedia.pub/entry/39121
Oliveira, Ellen Gonçalves De, et al. "Phycodnaviridae as Giant Viruses of Interest for Biotechnology." Encyclopedia. Web. 22 December, 2022.
Copy Citation
Giant viruses of the phylum Nucleocytoviricota infect protists (i.e., algae and amoebae) and have complex genomes, reaching up to 2.7 Mb in length and encoding hundreds of genes. Different giant viruses have robust metabolic machinery, especially those in the Phycodnaviridae families. The Phycodnaviridae family includes viruses with biochemical and genetic peculiarities, such as DNA error correction and post-replicative processing, that infect eukaryotic algae from freshwater or marine environments.
giant viruses
phycodnaviridae
biotechnology
1. Introduction
The global demand and trade of industrial enzymes are continuously growing, and they are estimated to reach $7.0 billion USD in the next few years [1]. In this scenario, great importance is given to microbial enzymes, presenting several advantages, such as high yields, activity, and reproducibility, in addition to economic production, exponential growth, use of cheap platforms, and easy optimization. Many industrial processes demand enzymes, such as producing food, pharmaceutical products, detergents, and textiles. In this context, recombinant gene technology, protein engineering, and directed evolution have revolutionized enzyme manufacturing and this industry. Enzymes with a hydrolytic activity are used in the degradation processes of various natural substances and are extensively applied in industry. Proteases, essential enzymes for the detergent and dairy industries, are also widely used. Those enrolled in carbohydrate metabolism, including amylases and cellulases, are extensively used in the textile, detergent, and food industries [2]. Approximately 60% of industrial enzymes come from fungi, 24% from bacteria, 4% from yeasts, and most of the 12% remaining are obtained from plants and animals [1][2]. However, although viruses represent a small portion of these enzymes, studies about their potential have been growing in the last 30 years (Figure 1). They have a complex geometric structure and highly efficient genetic machinery, and, beyond that, they are sources of unique enzymes with great biotechnological potential [3].
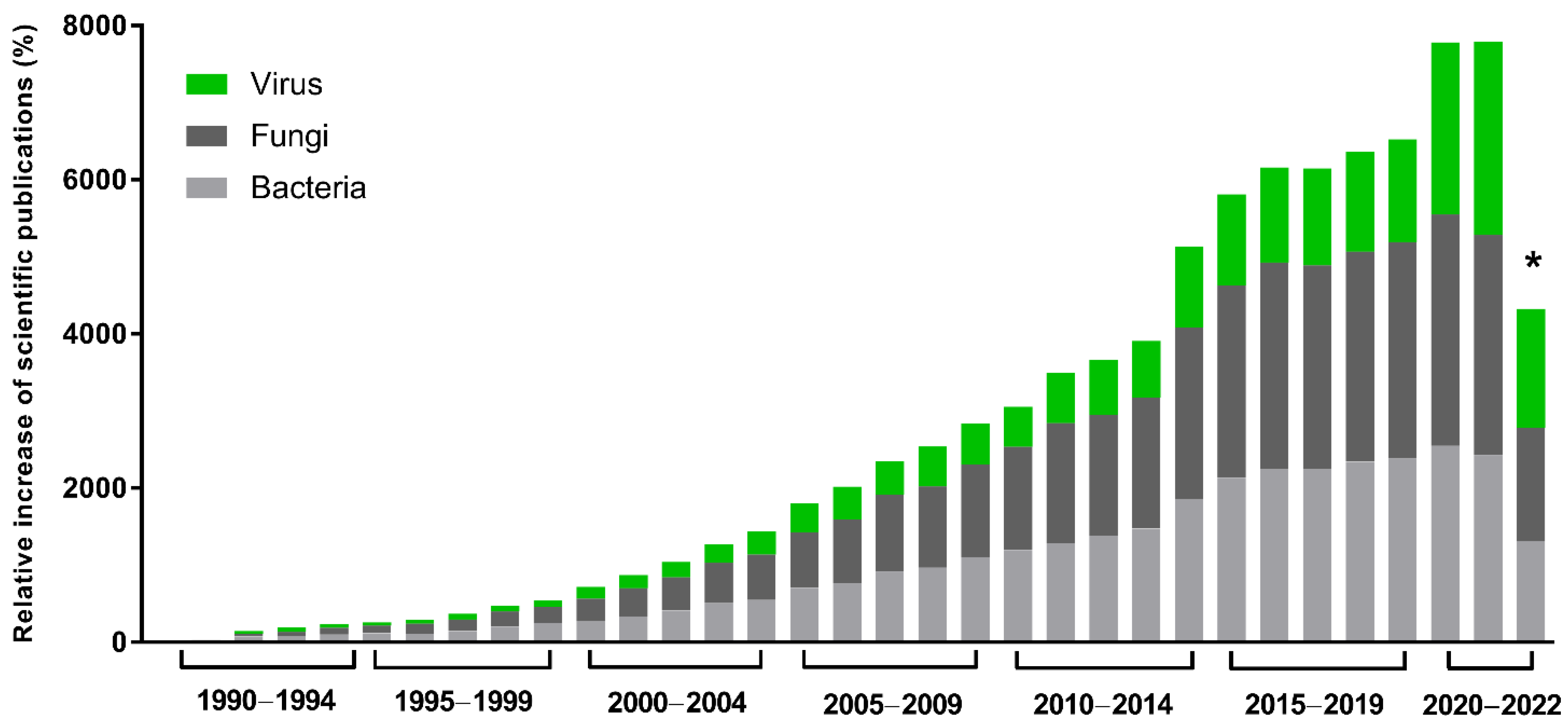
Figure 1. Scientific interest in microbial enzymes in biotechnology. Relative increase in scientific publications since 1990. The numbers were obtained from the Pubmed database using the names of the microbial groups (viruses, fungi, or bacteria), plus enzymes plus biotechnology. A total of 112,240 results were obtained. Relative increase was calculated by comparing the number of publications in a given year with the publication number in 1990, the first year of the historical record. * Data as of 12 September 2022.
The discovery of reverse transcriptase in retroviruses by David Baltimore and Howard Temin in 1970 was a milestone for molecular and cancer biology and started our understanding about retrovirology [4]. Other viruses that have been exploited for biotechnology are bacteriophages, since they are easy to manipulate and have interesting enzymes for several applications, such as DNA polymerases, DNA ligases, and lytic enzymes [2][5]. These lytic enzymes have enormous potential for use as antimicrobials because they exhibit bactericidal effects, absence of resistance, and activity against persistent cells. These enzymes degrade peptidoglycans, have antimicrobial and anti-biofilm properties (e.g., endolysins), and can be applied in treatments of bacterial infections [5][6].
The concept that viruses carry only genes that support their viral replication and capsid production changed with the discovery of giant viruses, opening space for a new approach and understanding of their contribution to the evolution of life [7]. Differently from the other viral groups and, similarly to bacteria and prokaryotes, they carry large genomes, with a diversity of genes capable of coding for numerous proteins, including DNA repair and even metabolic enzymes [7][8]. This new approach to understanding not only enriches the primary refinement regarding these viruses and their hosts, but also the beginning of the potential of these organisms for several biotechnological purposes. These viruses were first discovered in the 1970s, infecting unicellular algae, and many different isolates have been identified since then [9][10][11]. With the discovery of mimiviruses in the early 2000s and other giant amoeba viruses in the following years, the group of so-called nucleocytoplasmic large DNA viruses (NCLDV), currently classified in the phylum Nucleocytoviricota, greatly expanded, and pushed forward the boundaries of the virosphere [12][13][14].
Genomic studies of giant viruses of protists raised many questions about their biology, ecology, origin, and evolution. In addition, the surprising amount of genes harbored by these viruses, with a considerable number of them encoding enzymes used as valuable tools in different sectors of the economy, open a new venue for important novelties originated from viruses to be explored and applied in the biotechnology field [15][16][17].
2. Phycodnaviridae: The First Family of Giant Viruses of Protists
The Phycodnaviridae family includes viruses with biochemical and genetic peculiarities, such as DNA error correction and post-replicative processing, that infect eukaryotic algae from freshwater or marine environments [18][19]. Phylogenetic analysis using DNA polymerase B sequences from members of this family showed that they have a common ancestor with other NCLDV, thus corroborating their classification in the phylum Nucleocytoviricota [14]. The family currently comprises six genera named Coccolithovirus, Phaeovirus, Prasinovirus, Raphidovirus, Prymnesiovirus, and Chlorovirus, that differ in terms of cycle type, host, genome topology, and gene content [20][21].
Although genes enrolled in lipid metabolism are not the most abundant functional category in giant viruses, they are strongly present in coccolithoviruses (Figure 2). Coccolithoviruses infect microalgae of the species Emiliania huxleyi, commonly found in marine sediments. Emiliania huxleyi virus 86 (EhV-86), one the first known coccolithoviruses, was observed in 1999, having a viral particle around 200 nm, covered by a lipid membrane and a linear genome of 407 kbp [22]. Curiously, a genomic characterization of the EhV-86 identified 472 coding sequencing (CDS) regions, but only 63 genes have a known function so far (Table 1). An amount of 10% of them encode enzymes involved in the biosynthesis of sphingolipids: sterol desaturase, serine palmitoyltransferase, lipid phosphate phosphatase, and two genes encoding desaturases (Figure 2).
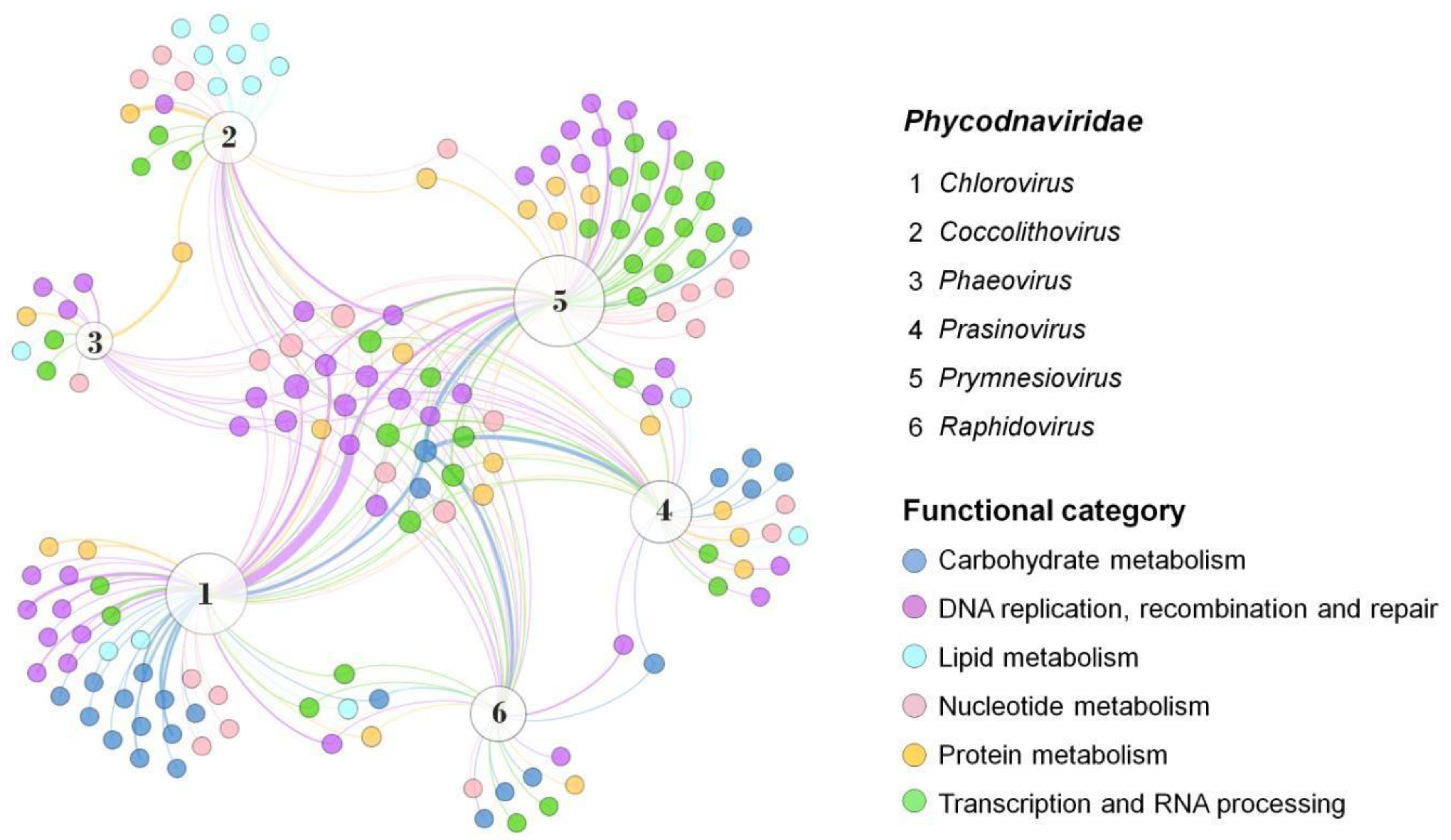
Figure 2. Presence and distribution of enzymes in the Phycodnaviridae family. Representatives of each genus were included and data on the diversity and abundance of enzymes grouped into different functional categories were obtained from genomic annotations publicly available on GenBank. A network graph was constructed using Gephi 0.9.7 using a force-based algorithm (ForceAtlas2), followed by a manual arrangement of nodes for better visualization. Node sizes are proportional to the degree of connection. The thickness of the edges is proportional to the number of genes of the same function in the genome of a virus. Virus representatives: (1) Paramecium bursaria chlorella virus 1; (2) Emiliania huxleyi virus 86; (3) Feldmannia species virus; (4) Ostreococcus tauri virus 5; (5) Phaeocystis globosa virus; (6) Heterosigma akashiwo virus 01.
Table 1. General genomic data of representatives of different groups of giant viruses of protists.
| Family * | Genus * | Virus | Genome Length (bp) | GC% | CDS | Unclassified CDS # | NCBI Accession |
|---|---|---|---|---|---|---|---|
| Phycodnaviridae | Chlorovirus | Paramecium bursaria Chlorella virus 1 | 330,611 | 40.0 | 416 | 368 | NC_000852.5 |
| Only Syngen Nebraska virus 5 | 327,147 | 42.4 | 357 | 204 | NC_032001.1 | ||
| PBCV CVB-1 | 319,457 | 44.3 | 339 | 214 | JX997160 | ||
| Acanthocystis turfacea chlorella virus 1 | 288,047 | 49.4 | 329 | 218 | NC_008724.1 | ||
| Coccolithovirus | Emiliania huxleyi virus 86 | 407,339 | 40.2 | 472 | 409 | AJ890364.1 | |
| Prasinovirus | Micromonas pusilla virus SP1 | 173,451 | 40.6 | 242 | 220 | NC_043129.1 | |
| Ostreococcus tauri virus OtV5 | 186,713 | 44.8 | 268 | 208 | NC_010191.2 | ||
| Raphidovirus | Heterosigma akashiwo virus 01 | 274,793 | 30.4 | 246 | 190 | NC_038553.1 | |
| Prymnesiovirus | Chrysochromulina parva virus | 437,255 | 25.1 | 45 | 28 | MH918795.1 | |
| Phaeovirus | Feldmannia species virus | 154,641 | 51.8 | 150 | 110 | NC_011183.1 | |
| Mimiviridae | Mimivirus | Acanthamoeba polyphaga mimivirus | 1,181,549 | 27.9 | 985 | 537 | HQ336222.2 |
| Moumouvirus australiensis | 1,098,000 | 25.1 | 899 | 438 | MG807320.1 | ||
| Megavirus chilensis | 1,246,130 | 25.3 | 1126 | 610 | NC_016072.1 | ||
| Tupanvirus | Tupanvirus soda lake | 1,516,267 | 29.0 | 1359 | 914 | KY523104.2 | |
| Tupanvirus deep ocean | 1,439,510 | 29.4 | 1276 | 845 | MF405918.2 | ||
| Cafeteriavirus | Cafeteria roenbergensis virus | 617,453 | 23.3 | 544 | 376 | NC_014637.1 | |
| Chlorella virus XW01 | 407,612 | 21.9 | 200 | 90 | OL828820.1 | ||
| Klosneuvirus | Klosneuvirus | 1,573,080 | 28.6 | 1545 | 1017 | KY684108.1 | |
| Fadolivirus | 1,573,504 | 27.1 | 1428 | 674 | MT418680.1 | ||
| Yasminevirus | 1,991,922 | 40.4 | 1434 | 926 | UPSH01000001.1 | ||
| Mesomimivirus | Bodo saltans virus | 1,385,870 | 25.3 | 1207 | 683 | MF782455.1 | |
| Tetraselmis virus 1 | 668,031 | 41.2 | 653 | 461 | KY322437.1 | ||
| Aureococcus anophagefferens virus | 370,920 | 28.9 | 384 | 309 | OM876856.1 | ||
| Marseilleviridae | Marseillevirus | Marseillevirus T19 | 368,454 | 44.7 | 428 | 273 | NC_013756.1 |
| Noumeavirus | 376,207 | 42.9 | 452 | 300 | NC_033775.1 | ||
| Tunisvirus | 380,011 | 43.0 | 484 | 355 | NC_038511.1 | ||
| Brazilian Marseillevirus | 362,276 | 43.3 | 491 | 347 | NC_029692.1 | ||
| Pithoviridae | Pithovirus | Pithovirus sibericum | 610,033 | 35.8 | 467 | 339 | NC_023423.1 |
| Cedratvirus | Cedratvirus A11 | 589,068 | 42.7 | 574 | 330 | NC_032108.1 | |
| Brazilian cedratvirus | 460,038 | 42.9 | 533 | 325 | LT994651.1 | ||
| Orpheoviridae | Orpheovirus | Orpheovirus IHUMI | 1,473,573 | 25.0 | 1199 | 753 | NC_036594.1 |
| Pandoraviridae | Pandoravirus | Pandoravirus salinus | 2,476,870 | 61.7 | 1430 | 853 | NC_022098.1 |
| Pandoravirus dulcis | 1,908,520 | 63.7 | 1070 | 748 | NC_021858.1 | ||
| Pandoravirus massiliensis | 1,593,060 | 60.1 | 1269 | 1003 | MZ384240.1 | ||
| Pandoravirus neocaledonia | 2,003,190 | 60.6 | 1081 | 709 | NC_037666.1 | ||
| Molliviridae | Mollivirus | Mollivirus sibericum | 651,523 | 60.1 | 523 | 424 | NC_027867.1 |
| Mollivirus kamchatka | 648,864 | 60.1 | 504 | 428 | MN812837.1 | ||
| Medusaviridae | Acanthamoeba castellanii medusavirus | 381,277 | 61.7 | 470 | 358 | AP018495.1 | |
| Faustoviridae | Faustovirus | Faustovirus E12 | 466,265 | 36.2 | 492 | 404 | KJ614390.1 |
| Faustovirus D3 | 465,956 | 37.7 | 485 | 423 | KU556803.1 | ||
| Pacmanvirus | Pacman A23 | 395,405 | 33.6 | 465 | 362 | LT706986.1 | |
| Kaumoebavirus | Kaumoebavirus Sc | 350,731 | 43.7 | 429 | 391 | NC_034249.1 |
* Only italicized taxa are currently defined by International Committee on Taxonomy of Viruses (ICTV). # Genes with unknown functions based on NCVOG functional categories.
Such enzymes are involved in the synthesis of ceramide, which induces apoptosis of the infected cell [23]. Although the mechanism of apoptosis has already been observed in other viruses, no genes related to the synthesis of sphingolipids were found in their genomes, making these genes exclusive to coccolithoviruses [24][25][26]. In addition, a proteome analysis showed that these enzymes enrolled in sphingolipid biosynthesis are present as early-class proteins, suggesting that they could be functional and also play an important role in initial infection [27]. Besides that, this highlights that these genes are not only carried inside the viral particles, but that they are also translated into functional proteins in the host and can be explored as biotechnological enzymes. Sphingolipids are molecules found in eukaryotes and prokaryotes and perform structural, signaling, and biochemical functions. They have been mentioned as a potent food supplement, and as a cosmetic, as they prevent skin infection and inhibit bacteria and fungi proliferation [28][29].
A comparative study performed by Nissimov and colleagues showed the presence of 25 to 29 CDS from other isolated viruses (EhV-201, EhV-207, and EhV-208) identical to sequences present in the EhV-86 genome. The predicted enzymes found were methyltransferases, glycosyltransferases, and RNase, and the majority of non-shared proteins, considered hypothetical ones, have unknown functions. On the other hand, the EhV-84 isolate showed many proteins (231 CDS) with identical homology with EhV-86 proteins [30]. With a few available genomes, it is clear that there is a vast field to be explored, both for obtaining more information about the viruses’ biology and ecology, and to be investigated for biotechnological purposes.
Phaeoviruses infect filamentous algae, most from the genus Ectocarpus and Feldmania, in subtropical environments, and they are the only phycodnaviruses known so far to infect more than one host. Genomic analysis of the Ectocarpus siliculosus virus-1 (EsV-1) revealed 231 CDS regions, where only 50% had determined functional characterization. Among these include genes involved in DNA synthesis, polysaccharide metabolism, histidine protein kinases, integration, and transposition [31]. Integrases catalyze site-specific DNA rearrangement, and transposases can bind in transposons on DNA and move small fragments along the genome. Both enzymes can be used for gene editing, gene therapy, and integrases are also studied as resistant markers [32][33]. A close relative is the Feldmania species virus (FsV), a phaeovirus associated with the brown filamentous algae Feldmania sp. This virus was considered the smallest giant viruses with a linear genome of 154 kbp and 150 CDS regions, of which only 25% had similarity with the database, such as those enrolled on DNA replication, transcription, nucleotide metabolism, and also lipid and protein metabolisms (Figure 2 and Figure 3) [34].
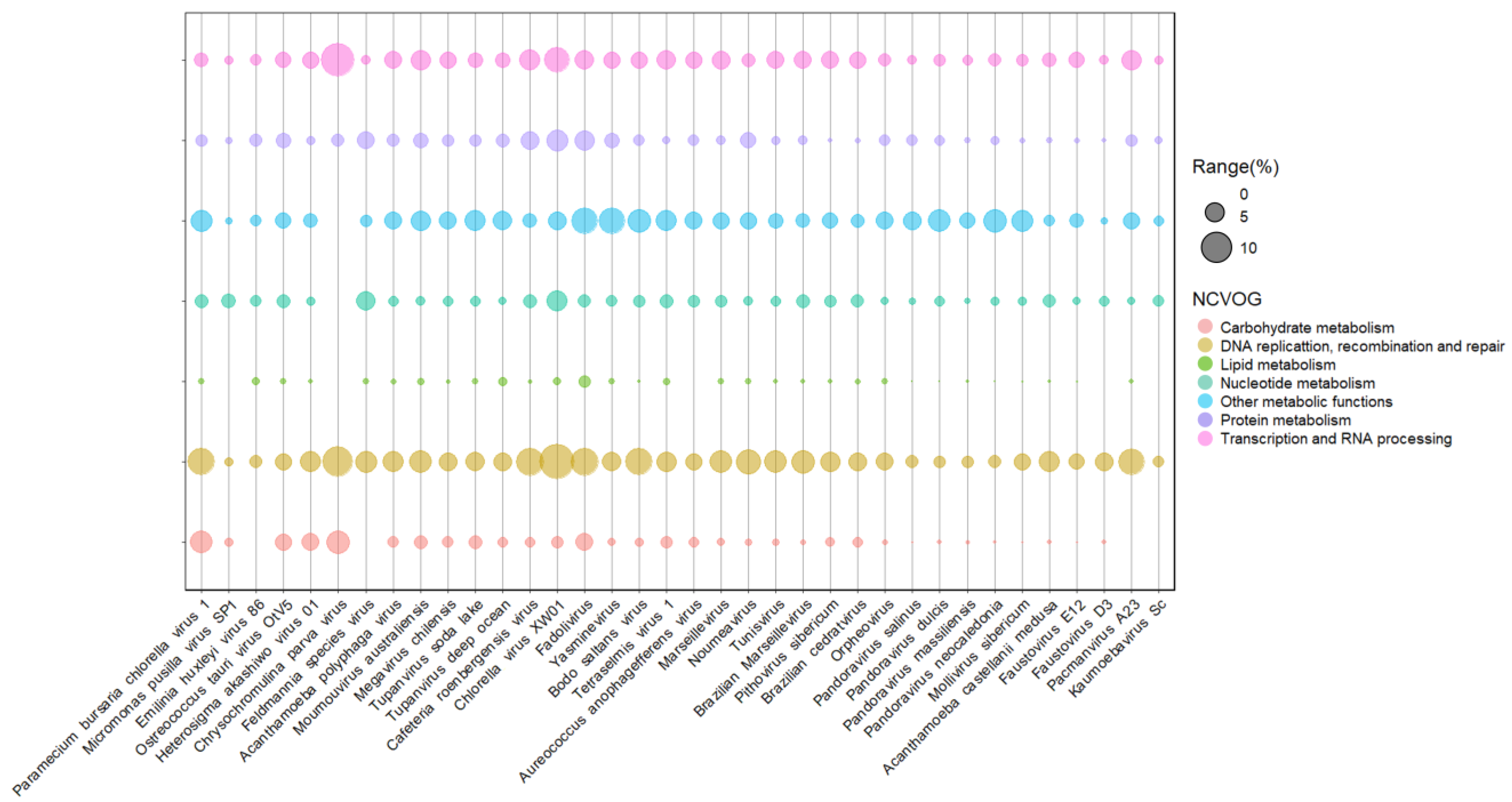
Figure 3. Presence and abundance of enzymes in giant viruses of protists. Bubble chart containing representatives of different viral families of the phylum nucleocytoviricota associated with algal and amoeba hosts. Bubble sizes are proportional to the number of enzymes represented as a gene percentage of a functional category in the virus genome. Data on enzyme diversity and abundance of each virus grouped into different functional categories were obtained from genomic annotations publicly available on GenBank and classified according to NCVOG categories.
Prasinoviruses infect prasinophytes, considered the smallest free-living photosynthetic eukaryotes [35]. Genomic analysis of the Osteococcus tauri virus (OtV-1) showed 232 CDS regions, where 31% showed functional similarity with previously described proteins, including methyltransferases and other enzymes involved in DNA, protein, and carbohydrate metabolism [33]. The Osteococcus tauri virus OtV-5 genome has 268 CDSs, and only 57% of the predicted proteins had a known function, including those involved in DNA replication and viral particle formation. Interestingly, some host-related genes were also found, including a proline dehydrogenase, related to cellular oxidation protective metabolism [36][37]. This virus has complex glycosylation machinery, with at least five glycosyltransferases and a galactosyltransferase, indicating relative independence of the host for glycosylating their own proteins (Figure 2). It’s worth noting that other giant viruses also have glycosylation machinery, with many proteins involved in carbohydrate modification and sugar production, which could be explored in the biotechnology industry [38][39].
Rhaphidoviruses have a wide variety of hosts. Among them is the single-celled seaweed bloom-forming Heterosigma akashiwo, which can form surface aggregations toxic to the environment [40]. The complete sequencing of the first virus strain infecting this alga (HaV53) was published in 2016, and genes related to DNA regulation, carbohydrate metabolism, signal transduction, and regulation of ubiquitin-related proteins were found. However, there is still a limited characterization of this genome [41]. Similar to other members of the Phycodnaviridae family, HaV01 has known glycosyltransferases that might be involved in viral protein glycosylation. In addition, proteins involved in transcription and RNA processing have also been identified, including a ribonuclease III and a mRNA-capping enzyme (Figure 2). Ribonuclease III can cleave double-strand RNA (dsRNA), an essential step in the maturation and decay of coding and non-coding RNAs. The first characterized ribonuclease III was from Escherichia coli, which is commercially available, and the enzyme is also present and well-conserved in plants, animals, fungi, and eukaryotic viruses [42]. The mRNA-capping enzyme is a complex that promotes the first modification of RNA polymerase II transcripts. In this context, this complex can regulate cap-dependent protein synthesis and act in the protein export mechanism [43]. Many types of mRNA-capping systems have also been described in viruses, such as influenza, orthomyxoviruses, alphaviruses, mimiviruses, and chloroviruses [43][44]. It is interesting to note that New England Biolabs Inc. has recently announced that the Faustovirus capping enzyme is commercially available, an enzyme that demonstrates increased capping efficiency across a variety of mRNA 5′ structures than previous enzymes [45].
Prymnesioviruses infect phytoplankton algae with high biomass formation, such as Phaeocystis globosa. Genomic analysis of the strain Phaetocistis globosa virus-16T (PgV-16T) showed 434 CDS regions with no phylogenetic proximity with the other viruses that infect algae, even though they are part of the Megaviridae clade. Seventy percent of its genome is similar to other large double-stranded DNA (dsDNA) viruses, with genes related to many processes, such as DNA replication and repair, including methyltransferases and transposases [46]. Seven coded genes seem unique in their genome among the group, which encode peculiar enzymes, such as phospholipase and asparagine synthetase homologs [46]. Phospholipases are responsible for hydrolyzing phospholipids into other lipids and are widely used in industrial food processes, while asparagine synthetase is a target related to the growth of human tumor cells. These prokaryote enzymes have also been characterized [47][48]. Compared to other phycodnaviruses, the difference between PgV-16T and these viruses’ genetic profile is clear, considering the functional clusters of genes (Figure 2). Such a difference corroborates previous data, pointing to this virus as a member of the Mimiviridae family [46]. Another member of this group is the Chrysochromulina brevifilum virus PW1, the only recognized species by ICTV so far [20]. A few viruses infecting Chrysochromulina sp. have been identified in the last years, and genome analysis of C. parva viruses suggested limited gene machinery compared to other phycodnaviruses (Figure 3).
The last-mentioned genus, Chlorovirus, was the first to be created, comprising the first virus associated with chlorella-like green algae, back in the late 1970s [49][50]. The first reported phycodnavirus, zoochlorella cell virus (ZCV), was isolated in the late 1970s in Japan from a Chlorella sp. that lives in symbiosis with the protozoan Paramecium bursaria. The ZCV was able to infect only zoochlorella recently separated from its symbiotic protozoan [9]. A few years later, viruses sharing many characteristics with ZCV were isolated from zoochlorella associated with Hydra viridis (HVCV-1 and HVCV-2) and also with Paramecium bursaria (PBCV-1), which would become the most studied algae viruses over the last decades [10][49][50].
References
- Fasim, A.; More, V.S.; More, S.S. Large-Scale Production of Enzymes for Biotechnology Uses. Curr. Opin. Biotechnol. 2021, 69, 68–76.
- Singh, R.; Kumar, M.; Mittal, A.; Mehta, P.K. Microbial Enzymes: Industrial Progress in 21st Century. 3 Biotech 2016, 6, 174.
- Varanda, C.; do Rosário Félix, M.; Campos, M.D.; Materatski, P. An Overview of the Application of Viruses to Biotechnology. Viruses 2021, 13, 2073.
- Coffin, J.M.; Fan, H. The Discovery of Reverse Transcriptase. Annu. Rev. Virol. 2016, 3, 29–51.
- Briers, Y. Phage Lytic Enzymes. Viruses 2019, 11, 113.
- Olsen, N.M.C.; Thiran, E.; Hasler, T.; Vanzieleghem, T.; Belibasakis, G.N.; Mahillon, J.; Loessner, M.J.; Schmelcher, M. Synergistic Removal of Static and Dynamic Staphylococcus aureus Biofilms by Combined Treatment with a Bacteriophage Endolysin and a Polysaccharide Depolymerase. Viruses 2018, 10, 438.
- Nasir, A.; Romero-Severson, E.; Claverie, J.-M. Investigating the Concept and Origin of Viruses. Trends Microbiol. 2020, 28, 959–967.
- Brahim Belhaouari, D.; Pires De Souza, G.A.; Lamb, D.C.; Kelly, S.L.; Goldstone, J.V.; Stegeman, J.J.; Colson, P.; La Scola, B.; Aherfi, S. Metabolic Arsenal of Giant Viruses: Host Hijack or Self-Use? eLife 2022, 11, e78674.
- Kawakami, H.; Kawakami, N. Behavior of a Virus in a Symbiotic System, Paramecium bursaria—Zoochlorella. J. Protozool. 1978, 25, 217–225.
- Van Etten, J.L.; Meints, R.H.; Burbank, D.E.; Kuczmarski, D.; Cuppels, D.A.; Lane, L.C. Isolation and Characterization of a Virus from the Intracellular Green Alga Symbiotic with Hydra viridis. Virology 1981, 113, 704–711.
- Van Etten, J.L.; Agarkova, I.V.; Dunigan, D.D. Chloroviruses. Viruses 2020, 12, 20.
- La Scola, B.; Audic, S.; Robert, C.; Jungang, L.; de Lamballerie, X.; Drancourt, M.; Birtles, R.; Claverie, J.-M.; Raoult, D. A Giant Virus in Amoebae. Science 2003, 299, 2033.
- Colson, P.; Yutin, N.; Shabalina, S.A.; Robert, C.; Fournous, G.; La Scola, B.; Raoult, D.; Koonin, E.V. Viruses with More Than 1000 Genes: Mamavirus, a New Acanthamoeba polyphagamimivirus Strain, and Reannotation of Mimivirus Genes. Genome Biol. Evol. 2011, 3, 737–742.
- Koonin, E.V.; Dolja, V.V.; Krupovic, M.; Varsani, A.; Wolf, Y.I.; Yutin, N.; Zerbini, F.M.; Kuhn, J.H. Global Organization and Proposed Megataxonomy of the Virus World. Microbiol. Mol. Biol. Rev. 2020, 84, e00061-19.
- Schulz, F.; Abergel, C.; Woyke, T. Giant Virus Biology and Diversity in the Era of Genome-Resolved Metagenomics. Nat. Rev. Microbiol. 2022, 20, 721–736.
- Endo, H.; Blanc-Mathieu, R.; Li, Y.; Salazar, G.; Henry, N.; Labadie, K.; de Vargas, C.; Sullivan, M.B.; Bowler, C.; Wincker, P.; et al. Biogeography of Marine Giant Viruses Reveals Their Interplay with Eukaryotes and Ecological Functions. Nat. Ecol. Evol. 2020, 4, 1639–1649.
- Guglielmini, J.; Woo, A.C.; Krupovic, M.; Forterre, P.; Gaia, M. Diversification of Giant and Large Eukaryotic DsDNA Viruses Predated the Origin of Modern Eukaryotes. Proc. Natl. Acad. Sci. USA 2019, 116, 19585–19592.
- Kang, M.; Dunigan, D.D.; Van Etten, J.L. Chlorovirus: A Genus of Phycodnaviridae That Infects Certain Chlorella-like Green Algae. Mol. Plant Pathol. 2005, 6, 213–224.
- Van Etten, J.L.; Schuster, A.M.; Girton, L.; Burbank, D.E.; Swinton, D.; Hattman, S. DNA Methylation of Viruses Infecting a Eukaryotic Chlorella-like Green Alga. Nucleic Acids Res. 1985, 13, 3471–3478.
- ICTV. Current ICTV Taxonomy Release. Available online: https://ictv.global/taxonomy (accessed on 1 October 2022).
- Van Etten, J.L.; Dunigan, D.D.; Nagasaki, K.; Schroeder, D.C.; Grimsley, N.; Brussaard, C.P.D.; Nissimov, J.I. Phycodnaviruses (Phycodnaviridae). In Encyclopedia of Virology, 4th ed.; Bamford, D.H., Zuckerman, M., Eds.; Academic Press: Oxford, UK, 2021; pp. 687–695. ISBN 978-0-12-814516-6.
- Schroeder, D.C.; Oke, J.; Malin, G.; Wilson, W.H. Coccolithovirus (Phycodnaviridae): Characterisation of a New Large DsDNA Algal Virus That Infects Emiliana huxleyi. Arch. Virol. 2002, 147, 1685–1698.
- Wilson, W.H.; Schroeder, D.C.; Allen, M.J.; Holden, M.T.G.; Parkhill, J.; Barrell, B.G.; Churcher, C.; Hamlin, N.; Mungall, K.; Norbertczak, H.; et al. Complete Genome Sequence and Lytic Phase Transcription Profile of a Coccolithovirus. Science 2005, 309, 1090–1092.
- Li, M.-L.; Stollar, V. Alphaviruses and Apoptosis. Int. Rev. Immunol. 2004, 23, 7–24.
- Allen, M.J.; Howard, J.A.; Lilley, K.S.; Wilson, W.H. Proteomic Analysis of the EhV-86 Virion. Proteome Sci. 2008, 6, 11.
- Nainu, F.; Shiratsuchi, A.; Nakanishi, Y. Induction of Apoptosis and Subsequent Phagocytosis of Virus-Infected Cells as an Antiviral Mechanism. Front. Immunol. 2017, 8, 1220.
- Ku, C.; Sheyn, U.; Sebé-Pedrós, A.; Ben-Dor, S.; Schatz, D.; Tanay, A.; Rosenwasser, S.; Vardi, A. A Single-Cell View on Alga-Virus Interactions Reveals Sequential Transcriptional Programs and Infection States. Sci. Adv. 2020, 6, eaba4137.
- Yunoki, K.; Kukino, O.; Nadachi, Y.; Fujino, T.; Ohnishi, M. Separation and Determination of Functional Complex Lipids from Chicken Skin. J. Am. Oil Chem. Soc. 2008, 85, 427–433.
- Miazek, K.; Lebecque, S.; Hamaidia, M.; Paul, A.; Danthine, S.; Willems, L.; Frédérich, M.; Pauw, E.D.; Deleu, M.; Richel, A.; et al. Sphingolipids: Promising Lipid-Class Molecules with Potential Applications for Industry. A Review. Biotechnol. Agron. Soc. Environ. 2016, 20, 321–336.
- Nissimov, J.I.; Worthy, C.A.; Rooks, P.; Napier, J.A.; Kimmance, S.A.; Henn, M.R.; Ogata, H.; Allen, M.J. Draft Genome Sequence of the Coccolithovirus emiliania Huxleyi Virus 202. J. Virol. 2012, 86, 2380–2381.
- Delaroque, N.; Müller, D.G.; Bothe, G.; Pohl, T.; Knippers, R.; Boland, W. The Complete DNA Sequence of the Ectocarpus siliculosus Virus EsV-1 Genome. Virology 2001, 287, 112–132.
- Merrick, C.A.; Zhao, J.; Rosser, S.J. Serine Integrases: Advancing Synthetic Biology. ACS Synth. Biol. 2018, 7, 299–310.
- Hickman, A.B.; Dyda, F. Mechanisms of DNA Transposition. Microbiol. Spectr. 2015, 3, 2.
- Schroeder, D.C.; Park, Y.; Yoon, H.-M.; Lee, Y.S.; Kang, S.W.; Meints, R.H.; Ivey, R.G.; Choi, T.-J. Genomic Analysis of the Smallest Giant Virus—Feldmannia Sp. Virus 158. Virology 2009, 384, 223–232.
- Weynberg, K.D.; Allen, M.J.; Ashelford, K.; Scanlan, D.J.; Wilson, W.H. From Small Hosts Come Big Viruses: The Complete Genome of a Second Ostreococcus tauri Virus, OtV-1. Environ. Microbiol. 2009, 11, 2821–2839.
- Derelle, E.; Ferraz, C.; Escande, M.-L.; Eychenié, S.; Cooke, R.; Piganeau, G.; Desdevises, Y.; Bellec, L.; Moreau, H.; Grimsley, N. Life-Cycle and Genome of OtV5, a Large DNA Virus of the Pelagic Marine Unicellular Green Alga Ostreococcus tauri. PLoS ONE 2008, 3, e2250.
- Zhang, L.; Becker, D.F. Connecting Proline Metabolism and Signaling Pathways in Plant Senescence. Front. Plant Sci. 2015, 6, 552.
- Piacente, F.; Gaglianone, M.; Laugieri, M.E.; Tonetti, M.G. The Autonomous Glycosylation of Large DNA Viruses. Int. J. Mol. Sci. 2015, 16, 29315–29328.
- Speciale, I.; Notaro, A.; Abergel, C.; Lanzetta, R.; Lowary, T.; Molinaro, A.; Tonetti, M.; Etten, J.; Castro, C. The Astounding World of Glycans from Giant Viruses. Chem. Rev. 2022, 122, 15717–15766.
- Mohamed, Z.A. Potentially Harmful Microalgae and Algal Blooms in the Red Sea: Current Knowledge and Research Needs. Mar. Environ. Res. 2018, 140, 234–242.
- Ogura, Y.; Hayashi, T.; Ueki, S. Complete Genome Sequence of a Phycodnavirus, Heterosigma akashiwo Virus Strain 53. Genome Announc. 2016, 4, e01279-16.
- Nicholson, A.W. Ribonuclease III Mechanisms of Double-Stranded RNA Cleavage. WIREs RNA 2014, 5, 31–48.
- Ramanathan, A.; Robb, G.B.; Chan, S.-H. MRNA Capping: Biological Functions and Applications. Nucleic Acids Res. 2016, 44, 7511–7526.
- Decroly, E.; Ferron, F.; Lescar, J.; Canard, B. Conventional and Unconventional Mechanisms for Capping Viral MRNA. Nat. Rev. Microbiol. 2012, 10, 51–65.
- NEB. New England Biolabs® Introduces Faustovirus Capping Enzyme, a Novel Enzymatic MRNA Capping Solution for MRNA Manufacturing. Available online: https://international.neb.com/about-neb/news-and-press-releases/new-england-biolabs-introduces-faustovirus-capping-enzyme-a-novel-enzymatic-mrna-capping-solution-for-mrna-manufacturing (accessed on 1 October 2022).
- Santini, S.; Jeudy, S.; Bartoli, J.; Poirot, O.; Lescot, M.; Abergel, C.; Barbe, V.; Wommack, K.E.; Noordeloos, A.A.M.; Brussaard, C.P.D.; et al. Genome of Phaeocystis globosa Virus PgV-16T Highlights the Common Ancestry of the Largest Known DNA Viruses Infecting Eukaryotes. Proc. Natl. Acad. Sci. USA 2013, 110, 10800–10805.
- De Maria, L.; Vind, J.; Oxenbøll, K.M.; Svendsen, A.; Patkar, S. Phospholipases and Their Industrial Applications. Appl. Microbiol. Biotechnol. 2007, 74, 290–300.
- Lomelino, C.L.; Andring, J.T.; McKenna, R.; Kilberg, M.S. Asparagine Synthetase: Function, Structure, and Role in Disease. J. Biol. Chem. 2017, 292, 19952–19958.
- Meints, R.H.; Van Etten, J.L.; Kuczmarski, D.; Lee, K.; Ang, B. Viral Infection of the Symbiotic Chlorella-like Alga Present in Hydra viridis. Virology 1981, 113, 698–703.
- Van Etten, J.L.; Meints, R.H.; Kuczmarski, D.; Burbank, D.E.; Lee, K. Viruses of Symbiotic Chlorella-like Algae Isolated from Paramecium bursaria and Hydra viridis. Proc. Natl. Acad. Sci. USA 1982, 79, 3867–3871.
More
Information
Subjects:
Virology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Revisions:
3 times
(View History)
Update Date:
27 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




