Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Shanni Hong | -- | 1971 | 2022-12-07 10:14:12 | | | |
| 2 | Vivi Li | Meta information modification | 1971 | 2022-12-08 03:21:34 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Hu, X.; Zhang, D.; Zeng, Z.; Huang, L.; Lin, X.; Hong, S. Aptamer-Based Probes for Cancer Diagnostics and Treatment. Encyclopedia. Available online: https://encyclopedia.pub/entry/38190 (accessed on 06 March 2026).
Hu X, Zhang D, Zeng Z, Huang L, Lin X, Hong S. Aptamer-Based Probes for Cancer Diagnostics and Treatment. Encyclopedia. Available at: https://encyclopedia.pub/entry/38190. Accessed March 06, 2026.
Hu, Xueqi, Dongdong Zhang, Zheng Zeng, Linjie Huang, Xiahui Lin, Shanni Hong. "Aptamer-Based Probes for Cancer Diagnostics and Treatment" Encyclopedia, https://encyclopedia.pub/entry/38190 (accessed March 06, 2026).
Hu, X., Zhang, D., Zeng, Z., Huang, L., Lin, X., & Hong, S. (2022, December 07). Aptamer-Based Probes for Cancer Diagnostics and Treatment. In Encyclopedia. https://encyclopedia.pub/entry/38190
Hu, Xueqi, et al. "Aptamer-Based Probes for Cancer Diagnostics and Treatment." Encyclopedia. Web. 07 December, 2022.
Copy Citation
Aptamers are single-stranded DNA or RNA oligomers that have the ability to generate unique and diverse tertiary structures that bind to cognate molecules with high specificity. In recent years, aptamer researches have witnessed a huge surge, owing to its unique properties, such as high specificity and binding affinity, low immunogenicity and toxicity, and simplicity of synthesis with negligible batch-to-batch variation. Aptamers may bind to targets, such as various cancer biomarkers, making them applicable for a wide range of cancer diagnosis and treatment. In cancer diagnostic applications, aptamers are used as molecular probes instead of antibodies.
aptamers
cancer diagnosis
cancer therapy
cell membrane biomarkers
extracellular biomarkers
intracellular biomarkers
1. Introduction
The fast expansion of new tools for molecular diagnostics and tumor-targeted therapy has raised demand for highly specific targeting ligands for biomarkers whose expression differ in tumor cells or tissues. Owing to the intrinsic heterogeneity of human tumors, targeting ligands can aid in the identification of tumor-specific biomarkers, resulting in enhanced diagnostics, predicted therapeutic response, and as a consequence, a decrease in unnecessary therapies for unresponsive oncological patients [1][2]. The common therapeutic options today are endocrine therapy, chemotherapy and targeted therapy utilizing antibodies that recognize intracellular or extracellular cancer biomarkers. Nevertheless, they are not always effective and may result in adverse effects and drug tolerance [3]. Hence, there is an ongoing need to develop novel and effective methods for cancer diagnosis and treatment.
Aptamers provide a very interesting substitute for previous widely utilized bioaffinity materials for diagnosis or therapeutic tools for drug delivery [4][5][6][7][8][9][10]. Aptamers consist of functional single-stranded DNA or RNA that can specifically bind to targets with high affinity. The synthesis of aptamers is based on an in vitro evolution process named “Systematic Evolution of Ligands by EXponential enrichment” (SELEX) [11][12]. This technology entails iterative screening of high-affinity nucleic acid ligands for a diverse range of targets, which includes small molecules, proteins, peptides, toxins, cells and tissues. Since its introduction in 1990, approximately 20 variants of original SELEX techniques have been developed [13][14]. The basic concept of the selection procedure is similar for both DNA and RNA oligonucleotides, and it consists of three crucial steps: (i) incubating the random oligonucleotide library with the selected targets, (ii) segmentation or isolation of bound oligonucleotides from library, and (iii) recovery and amplification of the target binding candidate for use in the next cycle [15]. Therefore, sequences with desired characteristics and especially tunable specificity and affinity can be amplified selectively from the initial pool of oligonucleotides.
Compared with traditional antibodies, aptamers have the advantages of higher stability over a wide range of temperatures, pH, and solvents, making them widely used. Furthermore, aptamers are easily modifiable with various functional moieties, and can be synthesized via chemical or enzymatic approaches, earning them the moniker of “chemical antibodies”. In addition, aptamers would not cause unnecessary immunological responses due to the absence of fragment crystallizable (Fc) regions, which would bind with Fc receptors expressed on the surface of immune cells. These properties give aptamers a low immunogenicity and good biocompatibility. Moreover, specific targeting of cancer biomarkers plays a vital role in aptamers-based probes. The affinity between aptamers and cancer biomarkers become the key of diagnosis, prognosis monitoring, and targeted therapy. Therefore, a variety of aptamers optimized for targeting cancers have been selected and applied, opening up a new way for developing personalized medicines. Based on these advantages, aptamers can be ideal candidates for in vivo biosensing, which is considered to be a simple and fast way to monitor cancer diagnosis, prognosis and therapeutic response. At the same time, aptamers can be used as carriers to deliver cargo to cancer cells or tissues, so they can also be used in cancer therapy. Numerous examples are currently being tested in clinical trials, such as Pegaptanib, a treatment for age-related macular degeneration (AMD), has been shown to be effective on the commercial market [16]. Therefore, in the past decade, a variety of aptamers for cancer biomarkers have been continuously optimized and developed, opening up new avenues for the development of personalized medicines.
As we know, cancer biomarkers are present in tumor tissues or serum that include DNA, mRNA, enzyme, metabolites, transcription factors, and cell surface receptor. In the past few decades, significant and substantial progress has been made in this field. Various promising techniques based on the specific recognition of intracellular biomarkers or biomarkers on the cancer cell surface have been developed. In this entry, researchers summarize the development and various applications of aptamer-based probes in cancer diagnostics and therapeutics according to the different location of targeted cancer biomarkers, as well as discuss current issues and future prospects in this field (Figure 1).
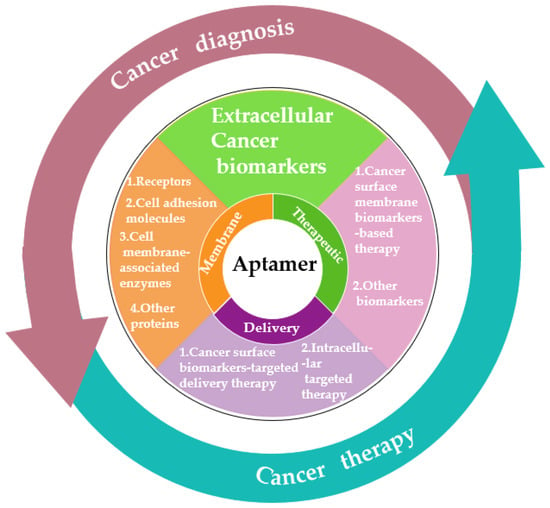
Figure 1. Schematic diagram of the application classification of aptamers in cancer diagnosis and treatment in the past decade.
2. Aptamer-Based Probes for Cancer Diagnosis
2.1. Cancer Surface Membrane Biomarkers
The cell surface membrane (PM) plays an important role in cell structure as a physical barrier that surrounds the cell and maintains the crucial boundaries between cytoplasm and the extracellular environment. The majority of PM mass is made up of proteins with critical specific functions. These proteins determine the methods of interaction between cells and its environment, including sending and receiving chemical signals, transportation metabolites, ions, or larger molecules, attached to neighboring cells and the extracellular matrix, etc. [17][18]. The mutation, deletion and overexpression of PM proteins are related to various pathological states of cancers. Currently, PM proteins have become the target for more than half of the approved drugs [19]. Some PM proteins (such as the human epidermal growth factor receptors 2 (HER2), mucin 1 (MUC1), and epithelial cell adhesion molecules) have been identified as crucial cancer biomarkers. Hence, a large number of specific aptamers have been developed against cell surface membrane biomarkers for basic research in physiology and biochemistry and also for diagnosis, monitoring and treatment of cancers.
2.2. Extracellular Cancer Biomarkers
The extracellular environment has a large impact on tumor and non-tumor tissues, especially in the concentrations of extracellular cancer biomarkers in body fluid [20]. Abnormal changes in cancer biomarker levels in body fluids are measuring standards of disease progression renewal. Therefore, the detection of these abnormal changes is often the key to realize early cancer diagnosis. Currently, the efforts are focused on liquid biopsy that relies on the presence of specific biomarkers in the body fluid of cancer patients. These cancer biomarkers are difficult to detect due to their low concentration levels in a high protein content medium. Enzyme-linked immunosorbent assays (ELISA) are the gold standard method for cancer biomarkers detection in body fluid, which rely on antibodies. Nevertheless, they suffer from some limitations, such as batch to batch variations during their production, and the challenging and cumbersome technique needed for generating specific monoclonal antibodies (especially against non-immunogenic molecules) [21][22]. For this reason, aptamers as novel receptors overcome these limitations owing to their unique characteristics of good stability, biocompatibility, safety, efficiency and non-immunogenicity. Herein, researchers summarized the various applications of aptamers in cancer diagnosis by introducing extracellular cancer biomarkers that have been detected in different body fluids.
Platelet-derived growth factor (PDGF), as one of serum components, has been proven to promote the proliferation of arterial smooth muscle cells [23]. The PDGF family is composed of four ligands: PDGF-A, B, C and D. PDGF-BB, a homodimer of PDGF-B, is an important cancer biomarker in diagnosis and recognition of cancers. During the past few years, aptamer-based recognition and detection of PDGF-BB for cancer diagnosis has been well-developed. Huang et al. developed an aptamer-modified AuNPs (Apt-GNPs) for the sensitive detection of PDGF-BB based on observing the changes in the color and extinction of the specific aptamer and GNPs by cause of aggregation [24]. This sensor was with a low detection limit of 35 nM and applied to protein analysis and cancer diagnosis. In another study, Tang et al. reported a strategy integrating rolling-circle amplification (RCA) and aptamer-based DNA enzyme-catalyzed colorimetric reaction for sensitive detection of PDGF-BB [25]. The PDGF-BB was recognized using primary aptamer-functionalized microbeads in a sandwich approach, and a secondary aptamer was attached to an RCA primer/circular template complex. The detection limit of this strategy was 8.2 fM. Zhang et al. constructed a AuNPs colorimetric sensor for detecting PDGF-BB by target-triggered strand displacement amplification system [26]. An obvious AuNPs color change can be observed when PDGF-BB concentration was as low as 4.0 nM. Interestingly, Ye et al. proposed a novel and simple aptamer-based one-two-three cascade DNA amplification surface-enhanced Raman scattering (SERS) strategy for the detection of PDGF-BB with the detection limit down to 0.42 pM [27]. Besides the strategies based on colorimetric and Raman scattering readout, fluorescent method is a simple and strong strategy because of its high sensitivity, rapid, simple and comparatively cost-less [2]. Taking advantages of fluorescent method, various aptamer-based fluorescent strategies have been developed for PDGF-BB detection. However, the labeling of aptamer with fluorophores and quenchers are the main limitations of the existing methods, which are time consuming and expensive. To overcome these limitations, Babu et al. reported an assay for label-free luminescent detecting PDGF by conjugating aptamer to hydrophobic Ru (II) complex as sensor system [28]. This method could detect the PDGF in a mixture of proteins, down to 0.8 pM. Wang et al. introduced a label-free and enzyme-free aptasensor for PDGF-BB quantification by using target-triggered hybridization chain reaction amplification and grapheme oxide (GO)-based selective fluorescence quenching with a detection limit of 1.25 pM [29]. Wang et al. reported another label-free fluorescent aptasensor for PDGF-BB detection by photo-induced electron transfer between DNA-AgNCs and G-quadruplex/hemin complexes. Binding of PDGF-BB to its aptamer caused a conformational change of DNA and the release of G-quadruplex sequence, which resulted in fluorescence change of the system [30]. In addition, Lin et al. developed a FRET based aptasensor using upconversion nanoparticles (UCNPs) as donor and AuNPs as acceptor for the PDGF-BB detection in blood serum of lymphoma patient with a low detection limit of 10 nM [31]. Compared to other biosensing methods, such as optical detection, the electrochemical aptasensors showed highly applicable and attractive for developing point-of-care cancer diagnosis tools owing to its advantages of disposability, accuracy, the ability to work with complex samples, easy control, rapid response, possible of usage for online control, etc. [32][33]. Recently, Jiang et al. developed a dual signal amplification for electrochemical aptasensing of PDGF-BB using hydroxyapatite nanoparticles (HAP-NPs) [34]. The phosphate group in both HAP-NPs and the aptamer reacted with molybdate to create a redox-active molybdophosphate precipitated on the surface of a glassy carbon electrode (GCE). When a voltage of 0.21 V (vs. Ag/AgCl) is applied, a current is generated whose intensity depended on the concentration of analyte. This work was applied to the determination of PDGF-BB in serum sample with a detection limit of 50 fg/mL.
3. Aptamer-Based Cancer Therapy
3.1. Aptamer as Therapeutic Agent
Aptamers are a group of potential therapeutic agents. They are known as “synthetic antibodies” because of their synthetic nucleic acid-based nature and excellent specificity and affinity for both protein and non-protein targets. The majority of aptamers utilized for therapeutic purposes are either chosen by in vivo selections using appropriate model systems or through in vitro selection utilizing a purified protein or receptor [35]. In clinical trials, therapeutic aptamers can be utilized as antagonists and agonists. Antagonist aptamers block or inhibit the integration of targets relevant to disease via protein–protein interaction or protein–receptor–ligand interaction [15]. Agonist aptamers can active the target receptors and also can be utilized as the carrier to carry the cargo to the target cells or tissues. Numerous studies have been reported that therapeutic aptamers applied for cancer therapy.
3.2. Aptamer as Delivery Agents
Aside from their utility as stand-alone therapeutics, aptamers can also serve as chaperones for another therapeutic. A variety of cell type-specific aptamers have been coupled with therapeutic drugs (such as siRNA, microRNA, anti-miR, therapeutic aptamer, chemotherapeutics, or toxins) or delivery vehicles (such as organic or inorganic nanocarriers) for cell type-specific delivery. Owing to the high specificity and affinity of aptamers, therapeutic compounds can be targeted to the desired cells or tissues, enhancing their local concentration and therapeutic efficacy.
References
- Cerchia, L.; Franciscis, V.D. Targeting cancer cells with nucleic acid aptamers. Trends Biotechnol. 2010, 28, 517–525.
- Hong, S.; Pawel, G.T.; Pei, R.; Lu, Y. Recent progress in developing fluorescent probes for imaging cell metabolites. Biomed. Mater. 2021, 16, 044108.
- Chen, H.; Zhang, W.; Zhu, G.; Xie, J.; Chen, X. Rethinking cancer nanotheranostics. Nat. Rev. Mater. 2017, 2, 17024.
- Sun, H.; Zu, Y. A highlight of recent advances in aptamer technology and its application. Molecules 2015, 20, 11959–11980.
- Mairal, T.; Özalp, V.C.; Sánchez, P.L.; Mir, M.; Katakis, I.; O’Sullivan, C.K. Aptamers: Molecular tools for analytical applications. Anal. Bioanal. Chem. 2008, 390, 989–1007.
- Hong, S.; Zhang, X.; Lake, R.J.; Pawel, G.T.; Guo, Z.; Pei, R.; Lu, Y. A photo-regulated aptamer sensor for spatiotemporally controlled monitoring of atp in the mitochondria of living cells. Chem. Sci. 2019, 11, 713–720.
- Hong, S.; Ding, P.; Luo, Y.; Gao, T.; Zhang, Y.; Pei, R. Aptamer-integrated a-gal liposomes as bispecific agents to trigger immune response for killing tumor cells. J. Biomed. Mater. Res. A 2019, 107, 1176–1183.
- Hong, S.; Sun, N.; Liu, M.; Wang, J.; Pei, R. Building a chimera of aptamer–antisense oligonucleotide for silencing galectin-1 gene. RSC Adv. 2016, 6, 112445–112450.
- Cheng, H.; Hong, S.; Wang, Z.; Sun, N.; Wang, T.; Zhang, Y.; Chen, H.; Pei, R. Self-assembled rnai nanoflowers via rolling circle transcription for aptamer-targeted sirna delivery. J. Mater. Chem. B 2018, 6, 4638–4644.
- Xu, L.; Hong, S.; Sun, N.; Wang, K.; Zhou, L.; Ji, L.; Pei, R. Berberine as a novel light-up i-motif fluorescence ligand and its application in designing molecular logic systems. Chem. Commun. 2016, 52, 179–182.
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822.
- Tuerk, C.; Gold, L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Sicence 1990, 249, 505–510.
- Radom, F.; Jurek, P.M.; Jacek Otlewski, M.P.M.; Jeleń, F. Aptamers: Molecules of great potential. Biotechnol. Adv. 2013, 31, 1260–1274.
- Aquino-Jarquin, G.; Toscano-Garibay, J.D. RNA aptamer evolution: Two decades of selection. Int. J. Mol. Sci. 2011, 12, 9155–9171.
- Kumar Kulabhusan, P.; Hussain, B.; Yüce, M. Current perspectives on aptamers as diagnostic tools and therapeutic agents. Pharmaceutics 2020, 12, 646.
- Zhou, B.; Wang, B. Pegaptanib for the treatment of age-related macular degeneration. Exp. Eye Res. 2006, 83, 615–619.
- Bretscher, M.S.; Raff, M.C. Mammalian plasma membranes. Nature 1975, 258, 43–49.
- Yıldırım, M.A.; Goh, K.; Cusick, M.E.; Barabási, A.L.; Vidal, M. Drug—Target network. Nat. Biotechnol. 2007, 25, 1119–1126.
- Cibiel, A.; Dupont, D.M.; Ducongé, F. Methods to identify aptamers against cell surface biomarkers. Pharmaceuticals 2011, 4, 1216–1235.
- Wu, L.; Qu, X. Cancer biomarker detection: Recent achievements and challenges. Chem. Soc. Rev. 2015, 44, 2963–2997.
- Lorenzo-Gómez, R.; Miranda-Castro, R.; de-los-Santos-Álvarez, N.; Lobo-Castañón, M.J. Electrochemical aptamer-based assays coupled to isothermal nucleic acid amplification techniques: New tools for cancer diagnosis. Curr. Opin. Electrochem. 2019, 14, 32–43.
- Toh, S.Y.; Citartan, M.; Gopinath, S.C.; Tang, T. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. Biosens. Bioelectron. 2015, 64, 392–403.
- Ross, R.; Glomset, J.; Kariya, B.; Harker, L. A platelet-dependent serum factor that stimulates the proliferation of arterial smooth muscle cells in vitro. Proc. Natl. Acad. Sci. USA 1974, 71, 1207–1210.
- Huang, C.; Huang, Y.; Cao, Z.; Tan, W.; Chang, H. Aptamer-modified gold nanoparticles for colorimetric determination of platelet-derived growth factors and their receptors. Anal. Chem. 2005, 77, 5735–5741.
- Tang, L.; Liu, Y.; Ali, M.M.; Kang, D.K.; Zhao, W.; Li, J. Colorimetric and ultrasensitive bioassay based on a dual-amplification system using aptamer and DNAzyme. Anal. Chem. 2012, 84, 4711–4717.
- Zhang, H.; Li, F.; Chen, H.; Ma, Y.; Qi, S.; Chen, X.; Zhou, L. AuNPs colorimetric sensor for detecting platelet-derived growth factor-BB based on isothermal target-triggering strand displacement amplification. Sens. Actuators B Chem. 2015, 207, 748–755.
- Ye, S.; Zhai, X.; Wu, Y.; Kuang, S. Dual-primer self-generation sers signal amplification assay for PDGF-BB using label-free aptamer. Biosens. Bioelectron. 2016, 79, 130–135.
- Babu, E.; Singaravadivel, S.; Manojkumar, P.; Krishnasamy, S.; Gnana kumar, G.; Rajagopal, S. Aptamer-based label-free detection of PDGF using ruthenium (II) complex as luminescent probe. Anal. Bioanal. Chem. 2013, 405, 6891–6895.
- Wang, X.; Jiang, A.; Hou, T.; Li, H.; Li, F. Enzyme-free and label-free fluorescence aptasensing strategy for highly sensitive detection of protein based on target-triggered hybridization chain reaction amplification. Biosens. Bioelectron. 2015, 70, 324–329.
- Wang, G.; Zhu, Y.; Chen, L.; Zhang, X. Photoinduced electron transfer (PET) based label-free aptasensor for platelet-derived growth factor-BB and its logic gate application. Biosens. Bioelectron. 2015, 63, 552–557.
- Lin, F.; Yin, B.; Li, C.; Deng, J.; Fan, X.; Yi, Y.; Liu, C.; Li, H.; Zhang, Y.; Yao, S. Fluorescence resonance energy transfer aptasensor for platelet-derived growth factor detection based on upconversion nanoparticles in 30% blood serum. Anal. Methods 2013, 5, 699–704.
- Meirinho, S.G.; Dias, L.G.; Peres, A.M.; Rodrigues, L.R. Voltammetric aptasensors for protein disease biomarkers detection: A review. Biotechnol. Adv. 2016, 34, 941–953.
- Razmi, N.; Baradaran, B.; Hejazi, M.; Hasanzadeh, M.; Mosafer, J.; Mokhtarzadeh, A.; de la Guardia, M. Recent advances on aptamer-based biosensors to detection of platelet-derived growth factor. Biosens. Bioelectron. 2018, 113, 58–71.
- Jiang, W.; Tian, D.; Zhang, L.; Guo, Q.; Cui, Y.; Yang, M. Dual signal amplification strategy for amperometric aptasensing using hydroxyapatite nanoparticles. Application to the sensitive detection of the cancer biomarker platelet-derived growth factor BB. Microchim. Acta 2017, 184, 4375–4381.
- Mi, J.; Liu, Y.; Rabbani, Z.N.; Yang, Z.; Urban, J.H.; Sullenger, B.A.; Clary, B.M. In vivo selection of tumor-targeting RNA motifs. Nat. Chem. Biol. 2010, 6, 22–24.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
924
Revisions:
2 times
(View History)
Update Date:
08 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No





