Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Rute G. Matos | -- | 9874 | 2022-12-05 13:05:35 | | | |
| 2 | Rute G. Matos | -8203 word(s) | 1671 | 2022-12-06 17:38:40 | | | | |
| 3 | Rita Xu | -19 word(s) | 1652 | 2022-12-07 03:13:46 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Costa, V.G.; Costa, S.M.; Saramago, M.; Cunha, M.V.; Arraiano, C.M.; Viegas, S.C.; Matos, R.G. RNA Technologies to Fight Human Pathogens. Encyclopedia. Available online: https://encyclopedia.pub/entry/38031 (accessed on 08 February 2026).
Costa VG, Costa SM, Saramago M, Cunha MV, Arraiano CM, Viegas SC, et al. RNA Technologies to Fight Human Pathogens. Encyclopedia. Available at: https://encyclopedia.pub/entry/38031. Accessed February 08, 2026.
Costa, Vanessa G., Susana M. Costa, Margarida Saramago, Marta V. Cunha, Cecília M. Arraiano, Sandra C. Viegas, Rute G. Matos. "RNA Technologies to Fight Human Pathogens" Encyclopedia, https://encyclopedia.pub/entry/38031 (accessed February 08, 2026).
Costa, V.G., Costa, S.M., Saramago, M., Cunha, M.V., Arraiano, C.M., Viegas, S.C., & Matos, R.G. (2022, December 05). RNA Technologies to Fight Human Pathogens. In Encyclopedia. https://encyclopedia.pub/entry/38031
Costa, Vanessa G., et al. "RNA Technologies to Fight Human Pathogens." Encyclopedia. Web. 05 December, 2022.
Copy Citation
A long scientific journey has led to prominent technological advances in the RNA field, and several new types of molecules have been discovered, from non-coding RNAs (ncRNAs) to riboswitches, small interfering RNAs (siRNAs) and CRISPR systems. Such findings, together with the recognition of the advantages of RNA in terms of its functional performance, have attracted the attention of synthetic biologists to create potent RNA-based tools for biotechnological and medical applications.
ribonucleases
small non-coding RNAs (ncRNAs)
virulence
RNA metabolism
1. Introduction
A crucial characteristic of the prokaryotic world is its rapid ability to adjust to a changing environment. In the case of pathogenic organisms, it is also essential that they overcome the host immune system. This implies an extensive and prompt re-adjustment of the gene expression by complex regulatory networks, in which RNA metabolism has a crucial role. In fact, RNA is much more than a messenger as it is able to dynamically coordinate and instruct cellular functions, and it has also emerged as an important feature to be considered for the pathogenesis of microorganisms.
Bacterial infections are associated with a high rate of human morbidity and mortality, and bacterial resistance to antibiotics is an escalating problem worldwide. The widespread use of conventional antibiotics has favored the apperance of drug-resistant pathogens, and there is a growing need for the development of novel antibacterial strategies. The idea of directly targeting RNA is emerging as a new frontier in drug discovery studies, with the ultimate goal of expanding the antibiotic arsenal. The differences between the molecular machinery that governs bacterial and eukaryotic RNA metabolism are fundamental to identify in order to take advantage of this attractive drug target.
The stability of messenger RNA relies on several features and involves numerous players, with ribonucleases (RNases) being among the most important ones. These enzymes are ubiquitous and can perform the RNA degradation alone or in multiprotein complexes. The diversity of RNA molecules with regulatory roles is better understood now, including a wide range of small non-coding RNAs (ncRNAs) and the natural RNA interference (RNAi), CRISPR/Cas (Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated protein) and ERASE (Endogenous Reverse Transcriptase/RNase H-mediated Antiviral System) systems. The ERASE system [1] is a DNA-mediated RNA cleavage mechanism that is parallel to the RNA-guided DNA cleavage of the CRISPR/Cas system and the RNA-guided RNA cleavage of the RNAi pathway. The prospect of using this system to fight pathogenic infections has not yet been explored.
Altogether, recent advances in RNA studies at a global scale have given researchers a vast amount of information about the role of the RNA regulation of pathogens. In this context, synthetic biology (SynBio) is emerging as a field that is focused on engineering biomolecular systems for a variety of applications. SynBio devices contribute not only to improve the understanding of the disease mechanisms, but also provide novel diagnostic tools. The developments in this field have created strategies for pathogen characterization, cancer treatment, vaccine development, microbiome engineering, cell therapy, regenerative medicine and the production of new and more affordable drugs. Additionally, the targeting of regulatory RNA-based interactions has broadened the SynBio applications in antimicrobial therapeutics. The inherent modularity and compatibility of RNA-based control components enables them to be independently optimized or exchanged, thus expanding their applications.
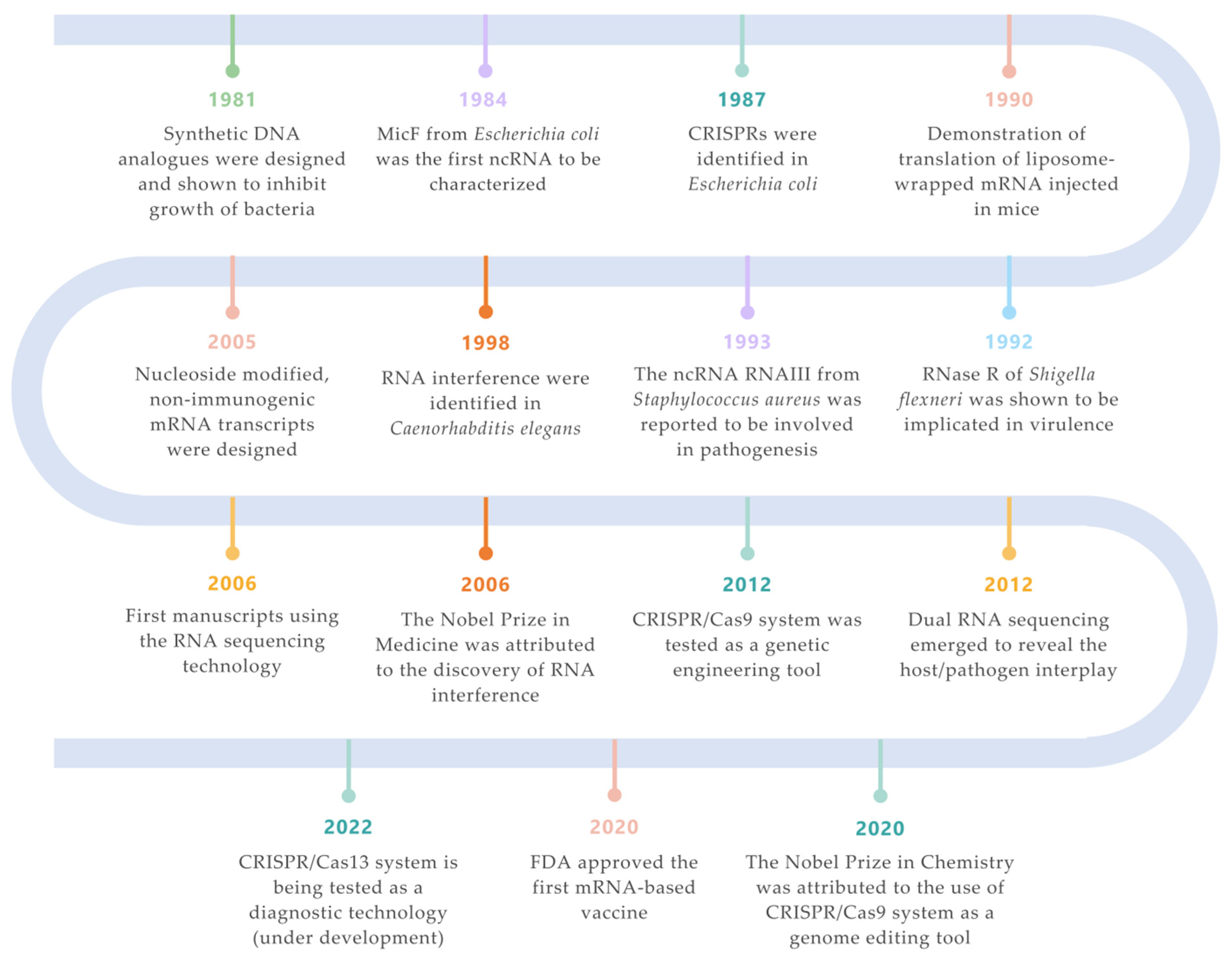
Figure 1. Landmarks on RNA technologies. An overview of the most relevant achievements and pioneer experiments around the RNA molecule. The timeline is color-coded for each field (green for synthetic biology; orange for siRNAs; purple for ncRNAs; turquoise for CRISPR/Cas systems; salmon for mRNA vaccines; yellow for RNA-seq technologies; blue for ribonucleases) [2][3][4][5][6][7][8][9][10][11][12][13].
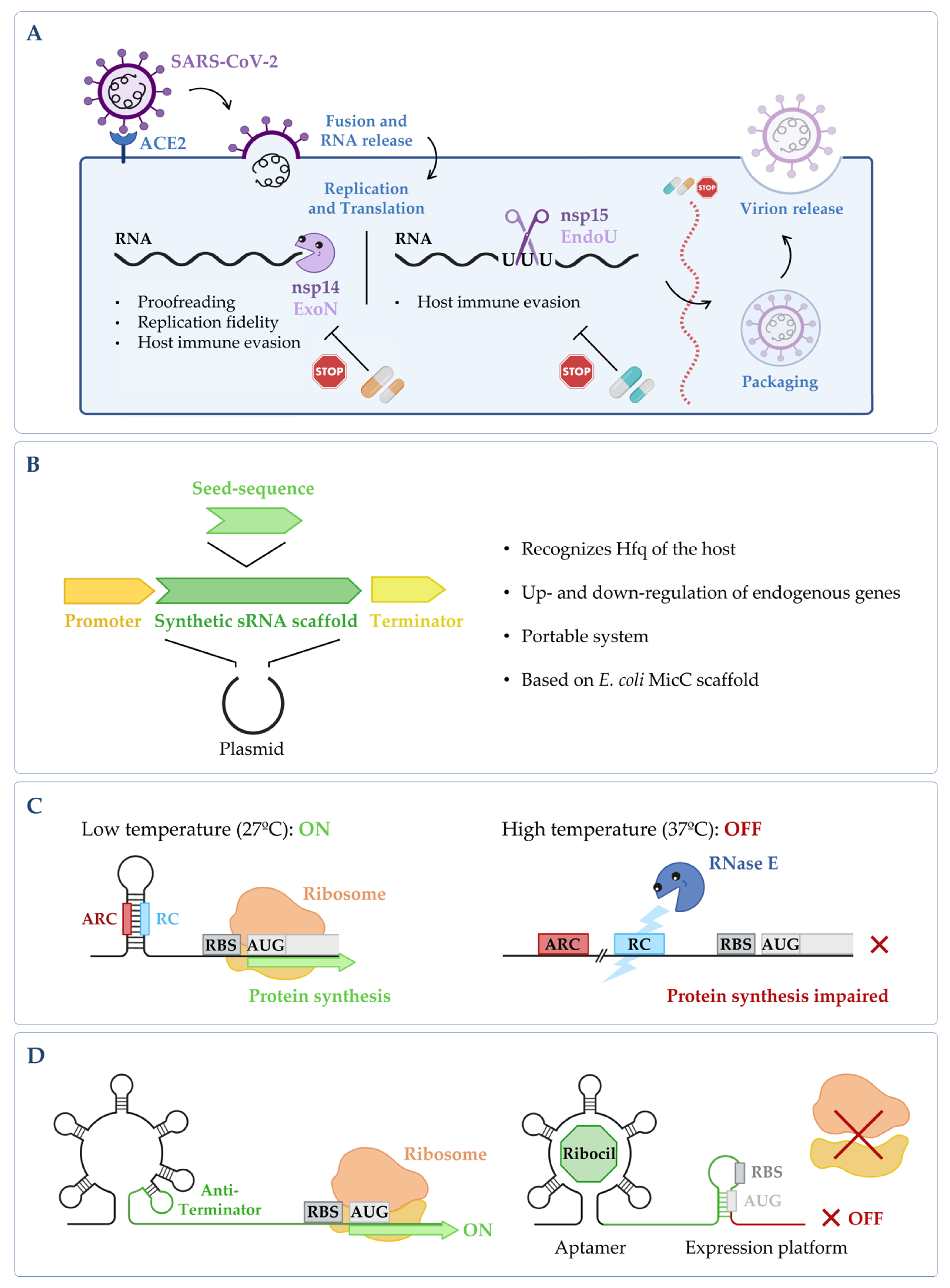
Figure 2. (A) Targeting fundamental viral proteins to inhibit SARS-CoV-2. The use of nsp14 and nsp15 ribonucleases as druggable targets may impair SARS-CoV-2 viral replication cycle, and therefore, it can be a good way to tackle infection. (B) Synthetic sRNA expression system. Vector containing the E. coli MicC scaffold, in which a customized seed sequence complementary to the endogenous target transcript is inserted [14]. (C) RNase E mediated thermoregulation. When temperature is low, the RNase E cleavage site (RC) is hybridized with the anti-RNase cleavage site (ARC) forming a hairpin, thus blocking the cleavage by RNase E and allowing gene expression to occur. When temperature increases, the RC is exposed, the mRNA is cleaved by RNase E, and the expression of the gene is impaired (RBS stands for ribosomal binding site and AUG for the initiation codon). This is adapted from [15]. (D) Regulation of the FMN riboswitch by Ribocil. FMN riboswitch in the absence of any compound; it presents a conformation that allows gene expression to occur. Upon binding to the riboswitch, Ribocil induces a rearrangement of its structure that sequesters the RBS, thus preventing translation; this is adapted from [16]. Figure created using BioRender.com (accessed on 11 November 2022).
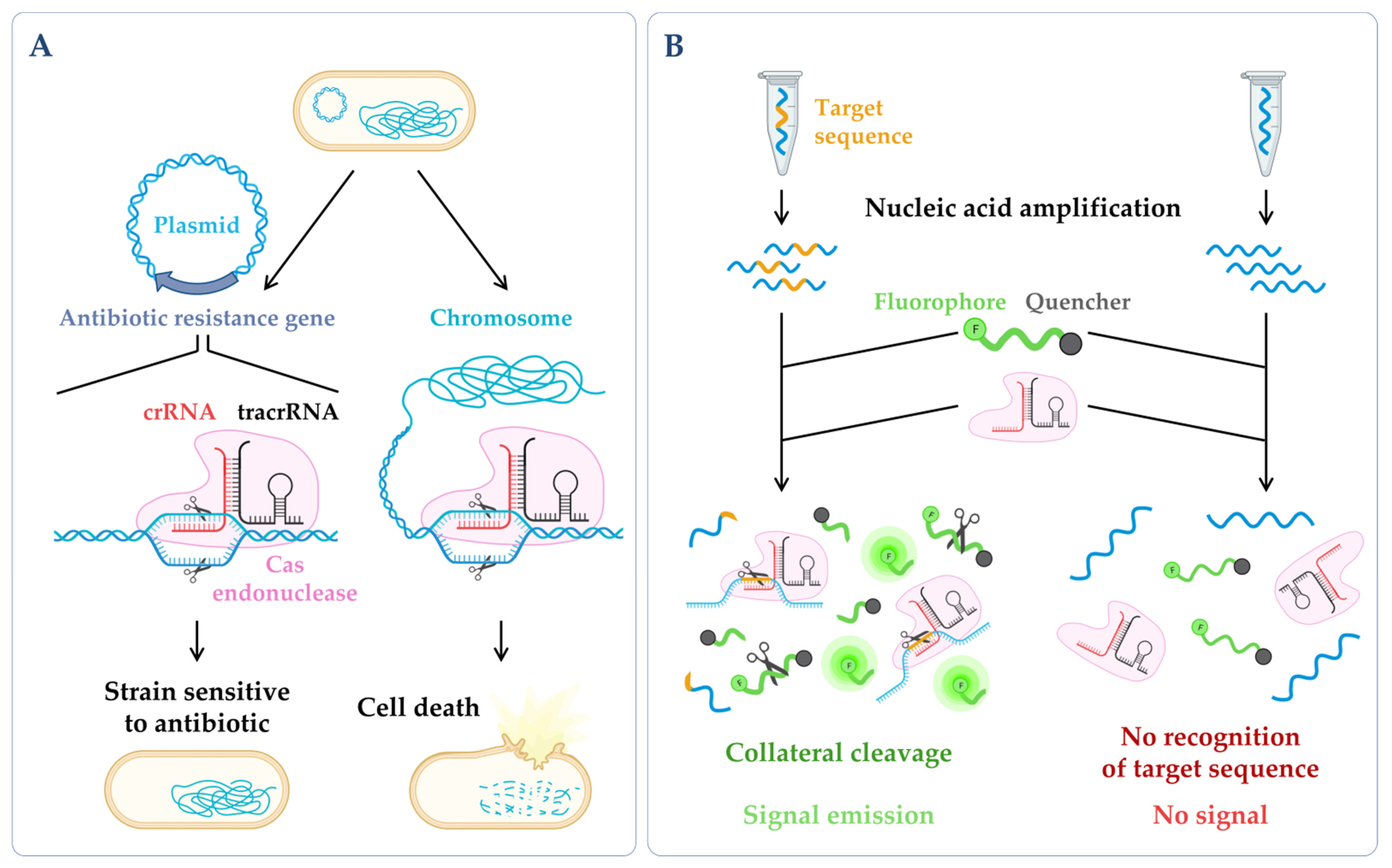
Figure 3. CRISPR technologies. (A) CRISPR-based antimicrobials. The system has been successfully tested through the directed degradation of the antibiotic resistance gene located in a plasmid (left side) leading to the recovery of the bacterial antibiotic sensitivity or the directed degradation of chromosomal genes, and consequently, cell death (bactericidal) [17]. (B) CRISPR-based diagnostics. When CRISPR effector proteins (Cas) recognize the specific target site, their collateral cleavage capability is triggered (this indiscriminate nucleic acid cleavage only happens when the crRNA finds its match). The addition of a reporter, that only releases the signal upon cleavage, enables the emission of a signal that can be easily detected [18]. Figure created using BioRender.com (accessed on 11 November 2022).
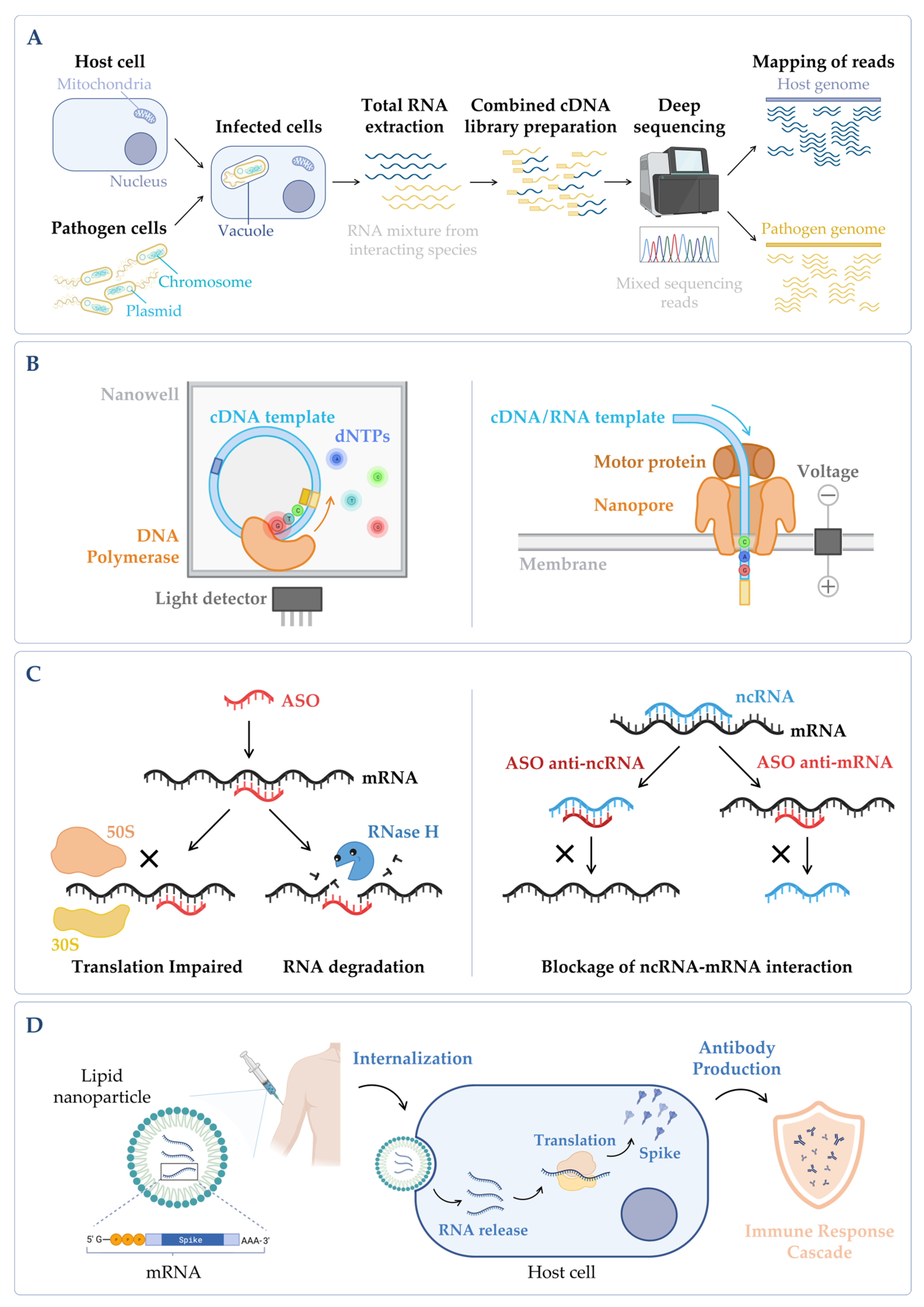
Figure 4. (A) Simplified workflow of a dual RNA-seq protocol. Host cells are infected in vitro with pathogen cells, lysed and total RNA is extracted. The sequencing library is prepared, and sequencing is performed in a NGS platform, obtaining simultaneously the results for both species. During bioinformatic data analysis, after quality control and data cleaning, the reads from the host and the pathogen are separated in silico in the mapping step. Annotation and quantification are carried out independently for each species, allowing to analyze host and pathogen differential gene expression in parallel, as well as to predict functional correlations between species [19]. (B) Main categories of third-generation sequencing (TGS). (Left panel) Single-molecule real-time (SMRT) sequencing—Sequence is determined through emission of fluorescence due to the incorporation of a fluorescently labelled deoxyribonucleotide (dNTP) by the DNA polymerase in the nascent complementary strand of the cDNA template molecule. The DNA polymerase is anchored to the bottom of a nanowell. (Right panel) Nanopore sequencing—Sequence is obtained without imaging. The template nucleic acid is bound to a motor protein which takes the molecule to a protein nanopore. When the template molecule is translocated through the pore, each nucleotide with its own modifications produces a characteristic current shift that is recorded. Unlike the other methods, direct RNA-seq uses an RNA molecule as template [20]. (C) Antisense oligonucleotides (ASOs) mechanism. (Left panel) General mechanism of ASOs activity. The oligonucleotide binds to the complementary RNA, impairing ribosome progression and/or causing transcript cleavage of a target duplex of mRNA/ASO by RNase H. (Right panel) Targeting of ncRNA–mRNA interaction. In this case, the ASO can be designed to mimic the ncRNA and block its binding to the mRNA (anti-mRNA ASO) or mimic the mRNA sequence to sequester the ncRNA (anti-ncRNA ASO) [21]. (D) mRNA vaccines mechanism. The nucleoside-modified mRNA containing the coding sequence of the protein of interest (SARS-CoV-2 Spike protein) is encapsulated in a lipid nanoparticle (LNP). Upon human vaccination, the LNP is internalized, and the mRNA coding sequence is recognized by the host translation machinery, leading to the production of Spike proteins. This will induce the production of specific antibodies by the host immune system, inducing an immune response cascade [22]. Figure created using BioRender.com (accessed on 11 November 2022).
2. Conclusions
RNA is back in the spotlight. The diverse role of RNA in all biological processes, together with the recognition of its important functional properties, have led to its exploitation in a wide range of biotechnological and medical applications. A great contributor to this change of perspective was the validation of the mRNA vaccines at an unpredictable scale and speed at which they fought against the COVID-19 pandemic. As they are natural molecules, RNAs present, in general, low toxicity and immunogenicity. The use of RNA elements presents advantages such as its independent control, tunability, composability and portability which empower their use as genetic tools. However, the advances in the application of these RNA tools have been limited by the rhythm of the progression of the technological advances which have enabled the characterization of new molecules and biological mechanisms. Built upon decades of scientific research, robust and prompt RNA technologies have now emerged, highlighting the importance of fundamental and applied research. For instance, researchers have testified in recent years, a fast discovery for the new classes of RNA molecules and molecular mechanisms that have transformed the comprehension of RNA metabolism. This research puts together the major discoveries regarding the connection between RNA metabolism and pathogenesis, and how this knowledge has been used to create new strategies to fight microbial pathogenicity. Antibiotic resistance is a serious problem that requires the creation of alternative therapeutics. As such, several RNA tools have surfaced as alternatives to control the virulence of pathogenic bacteria, namely, using synthetic non-coding RNAs, antisense antimicrobial therapeutics with antisense oligonucleotides (ASOs) or CRISPR–Cas antimicrobials. The application of these tools in prokaryotic organisms has been limited by different obstacles. In the case of ASOs, their use has been mostly limited by the development of delivery systems to improve their uptake by the bacterial cells. The same has happened with the CRISPR–Cas tools, and despite the new delivery systems which have been used with success, more research is needed to assure their safety and effectiveness. In the case of the mRNA vaccines, their implementation was possible thanks to the curiosity-driven studies of lipids and experiments with synthetic mRNA. The establishment of mRNA vaccines seems promising because of the speed with which they can be developed and produced, and their flexibility and adaptability to variants.
References
- Wu, J.; Wu, C.; Xing, F.; Cao, L.; Zeng, W.; Guo, L.; Li, P.; Zhong, Y.; Jiang, H.; Luo, M.; et al. Endogenous reverse transcriptase and RNase H-mediated antiviral mechanism in embryonic stem cells. Cell Res. 2021, 31, 998–1010.
- Tobe, T.; Sasakawa, C.; Okada, N.; Honma, Y.; Yoshikawa, M. Vacb, a novel chromosomal gene required for expression of virulence genes on the large plasmid of Shigella flexneri. J. Bacteriol. 1992, 174, 6359–6367.
- Jayaraman, K.; McParland, K.; Miller, P.; Ts’o, P.O. Selective inhibition of Escherichia coli protein synthesis and growth by nonionic oligonucleotides complementary to the 3′ end of 16s rRNA. Proc. Natl. Acad. Sci. USA 1981, 78, 1537–1541.
- Mizuno, T.; Chou, M.Y.; Inouye, M. A unique mechanism regulating gene expression: Translational inhibition by a complementary RNA transcript (micRNA). Proc. Natl. Acad. Sci. USA 1984, 81, 1966–1970.
- Ishino, Y.; Shinagawa, H.; Makino, K.; Amemura, M.; Nakata, A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J. Bacteriol. 1987, 169, 5429–5433.
- Wolff, J.A.; Malone, R.W.; Williams, P.; Chong, W.; Acsadi, G.; Jani, A.; Felgner, P.L. Direct gene transfer into mouse muscle in vivo. Science 1990, 247, 1465–1468.
- Novick, R.P.; Ross, H.F.; Projan, S.J.; Kornblum, J.; Kreiswirth, B.; Moghazeh, S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993, 12, 3967–3975.
- Kariko, K.; Buckstein, M.; Ni, H.; Weissman, D. Suppression of RNA recognition by toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 2005, 23, 165–175.
- Cheung, F.; Haas, B.J.; Goldberg, S.M.; May, G.D.; Xiao, Y.; Town, C.D. Sequencing Medicago truncatula expressed sequenced tags using 454 life sciences technology. BMC Genom. 2006, 7, 272.
- Bainbridge, M.N.; Warren, R.L.; Hirst, M.; Romanuik, T.; Zeng, T.; Go, A.; Delaney, A.; Griffith, M.; Hickenbotham, M.; Magrini, V.; et al. Analysis of the prostate cancer cell line lncap transcriptome using a sequencing-by-synthesis approach. BMC Genom. 2006, 7, 246.
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821.
- Westermann, A.J.; Gorski, S.A.; Vogel, J. Dual RNA-seq of pathogen and host. Nat. Rev. Microbiol. 2012, 10, 618–630.
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811.
- Apura, P.; Saramago, M.; Peregrina, A.; Viegas, S.C.; Carvalho, S.M.; Saraiva, L.M.; Arraiano, C.M.; Domingues, S. Tailor-made sRNAs: A plasmid tool to control the expression of target mRNAs in Pseudomonas putida. Plasmid 2020, 109, 102503.
- Hoynes-O’Connor, A.; Hinman, K.; Kirchner, L.; Moon, T.S. De novo design of heat-repressible RNA thermosensors in E. coli. Nucleic Acids Res. 2015, 43, 6166–6179.
- Wang, H.; Mann, P.A.; Xiao, L.; Gill, C.; Galgoci, A.M.; Howe, J.A.; Villafania, A.; Barbieri, C.M.; Malinverni, J.C.; Sher, X.; et al. Dual-targeting small-molecule inhibitors of the Staphylococcus aureus FMN riboswitch disrupt riboflavin homeostasis in an infectious setting. Cell Chem. Biol. 2017, 24, 576–588.e6.
- Kiga, K.; Tan, X.E.; Ibarra-Chavez, R.; Watanabe, S.; Aiba, Y.; Sato’o, Y.; Li, F.Y.; Sasahara, T.; Cui, B.; Kawauchi, M.; et al. Development of CRISPR-Cas13a-based antimicrobials capable of sequence-specific killing of target bacteria. Nat. Commun. 2020, 11, 2934.
- Gootenberg, J.S.; Abudayyeh, O.O.; Lee, J.W.; Essletzbichler, P.; Dy, A.J.; Joung, J.; Verdine, V.; Donghia, N.; Daringer, N.M.; Freije, C.A.; et al. Nucleic acid detection with CRISPR-Cas13a/c2c2. Science 2017, 356, 438–442.
- Westermann, A.J.; Forstner, K.U.; Amman, F.; Barquist, L.; Chao, Y.; Schulte, L.N.; Muller, L.; Reinhardt, R.; Stadler, P.F.; Vogel, J. Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 2016, 529, 496–501.
- Goodwin, S.; McPherson, J.D.; McCombie, W.R. Coming of age: Ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016, 17, 333–351.
- Henderson, C.A.; Vincent, H.A.; Callaghan, A.J. Reprogramming gene expression by targeting RNA-based interactions: A novel pipeline utilizing RNA array technology. ACS Synth. Biol. 2021, 10, 1847–1858.
- Walsh, E.E.; Frenck, R.W., Jr.; Falsey, A.R.; Kitchin, N.; Absalon, J.; Gurtman, A.; Lockhart, S.; Neuzil, K.; Mulligan, M.J.; Bailey, R.; et al. Safety and immunogenicity of two RNA-based COVID-19 vaccine candidates. N. Engl. J. Med. 2020, 383, 2439–2450.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
673
Revisions:
3 times
(View History)
Update Date:
07 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




