Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Katterine A. Salazar | -- | 3006 | 2022-11-30 21:04:40 | | | |
| 2 | Vivi Li | -1 word(s) | 3005 | 2022-12-01 02:53:35 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Oviedo, M.J.; Ramírez, E.; Cifuentes, M.; Farkas, C.; Mella, A.; Bertinat, R.; Gajardo, R.; Ferrada, L.; Jara, N.; Lima, I.D.; et al. IIIG9 in Cancer and Other Pathologies. Encyclopedia. Available online: https://encyclopedia.pub/entry/37404 (accessed on 07 February 2026).
Oviedo MJ, Ramírez E, Cifuentes M, Farkas C, Mella A, Bertinat R, et al. IIIG9 in Cancer and Other Pathologies. Encyclopedia. Available at: https://encyclopedia.pub/entry/37404. Accessed February 07, 2026.
Oviedo, María José, Eder Ramírez, Manuel Cifuentes, Carlos Farkas, Andy Mella, Romina Bertinat, Roberto Gajardo, Luciano Ferrada, Nery Jara, Isabelle De Lima, et al. "IIIG9 in Cancer and Other Pathologies" Encyclopedia, https://encyclopedia.pub/entry/37404 (accessed February 07, 2026).
Oviedo, M.J., Ramírez, E., Cifuentes, M., Farkas, C., Mella, A., Bertinat, R., Gajardo, R., Ferrada, L., Jara, N., Lima, I.D., Martínez, F., Nualart, F., & Salazar, K. (2022, November 30). IIIG9 in Cancer and Other Pathologies. In Encyclopedia. https://encyclopedia.pub/entry/37404
Oviedo, María José, et al. "IIIG9 in Cancer and Other Pathologies." Encyclopedia. Web. 30 November, 2022.
Copy Citation
The identification of new proteins that regulate the function of one of the main cellular phosphatases, protein phosphatase 1 (PP1), is essential to find possible pharmacological targets to alter phosphatase function in various cellular processes, including the initiation and development of multiple diseases. IIIG9 is a regulatory subunit of PP1 initially identified in highly polarized ciliated cells. In addition to its ciliary location in ependymal cells, researchers showed that IIIG9 has extraciliary functions that regulate the integrity of adherens junctions.
IIIG9
protein phosphatase 1
adherens junctions
ependymal cells
hydrocephaly
ciliopathies
ependymoma
1. Introduction
IIIG9 is a poorly characterized protein. This protein is encoded by the PPP1R32 gene, also known as C11orf66, FLJ32771, 4930579J09Rik, AU015816, and MGC144717. The human IIIG9 orphan gene (C11orf66) is located at the 11q12.2 locus, and its mRNA expression was first reported in 2002, when high expression was demonstrated in the ependymal wall of the fourth cerebral ventricle [1]. In rats, IIIG9 exists as two transcripts, IIIG9L (1491 bp) and IIIG9S (1355 bp), which leads to the emergence of 426 and 381 amino acid proteins, respectively. Both isoforms differ by 44 amino acids that are present in the N-terminal region of IIIG9L but absent in IIIG9S [1]. In humans, there are two IIIG9 isoforms: one of 425 amino acids (1532 bp) and the other of 405 amino acids (1472 bp) [2]. The short isoform does not have the sequence found between amino acids 230 to 249 (UniProt data), which are present in the long isoform. In mice, even when two transcripts are generated (1511 bp and 1515 bp), only a single protein of 427 amino acids is reported [3].
2. IIIG9 Is a Regulatory Subunit of Protein Phosphatase 1 (PP1)
IIIG9 was initially described as a protein of unknown function because it showed no significant homology to any known protein [1]. Additionally, according to the GCG Program ‘Motifs’ algorithm, it was not possible to identify any hydrophobic amino acid clusters indicative of signal sequences or transmembrane regions [1]. However, the rat IIIG9 protein sequence contains significant matches with sequences in other species such as the chicken and zebrafish [1]. Recently, with the aid of residue contact maps and artificial intelligence, it has become possible to generate a three-dimensional structure model of IIIG9 through the use of the C-I-ITASSER server [4][5]. After over 600 nanoseconds of molecular dynamics, IIIG9 reached a stable globular structure with the absence of large secondary structures and the presence of short alpha helices, in which the RKVHF sequence can be located. There are also a few beta-sheets at the level of the N-terminal region and a destructured region in the C-terminal region (Figure 1). The lack of secondary structures as intrinsically disordered proteins and the presence of the RKVHF sequence characterize more than 70% of PP1 regulatory proteins, which allow them to adopt highly flexible, transient conformations, thus promoting interaction with the protein surface of PP1 (AlphaFold: Protein Structure Database) [6][7][8]. The RKVHF sequence is a short, degenerate domain present between amino acids 129 and 133 (long isoform) or 85 and 89 (short isoform) in rat proteins or between amino acids 129 and 133 in both human and mouse isoforms [9] (Figure 1). Additionally, the interaction of IIIG9 with PP1 has been validated in vitro and then used in in silico screening reports for proteins that present an RVxF-type binding domain [9][10]. In conclusion, IIIG9 was identified as a candidate protein that interacts with PP1.
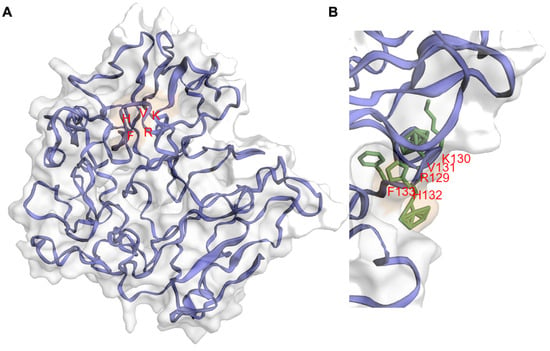
Figure 1. Molecular dynamics-refined model structure of human PPP1R32. (A) Cartoon representation of the best human PPP1R32 model according to the Z-score assessed by the ProSA webserver after 700 nanoseconds of molecular dynamics simulations. Protein structure, side chains, and surface patches were rendered with the EzMol server [11][12]. The model can reach a globular stable structure after 400 nanoseconds of molecular dynamics simulations. In the PPP1R32 protein model, the RKVHF motif (depicted with red letters) is accessible to the solvent. (B) Snapshot of RKVHF motif, depicting a histidine residue fully accessible to the solvent.
3. Regulation of PP1 by IIIG9
PP1 is a ubiquitous and highly conserved phosphatase, targeting about half of the proteins of eukaryotic cells. It is thought to cleave the ester connection of phosphates linked to serines or threonines [13]. In mammals, there are multiple PP1 catalytic subunits (α, β, and γ), and their locations and functions differ based on their interactions with more than 200 regulatory protein subunits [10][14]. In humans, three isoforms of PP1α have been described, including a 330 aa (1421 bp) canonical sequence and two isoforms produced by alternative splicing (both computationally predicted) of 286 (1289 bp) and 341 amino acids (1454 bp) (UniProt). The PP1β catalytic subunit has two transcripts of 4916 bp and 4925 bp, which lead to a single isoform of 327 amino acids. Finally, two alternative splicing isoforms have been described for PP1γ: PP1 gamma 1 (2552 bp) and PP1 gamma 2 (1521 bp), with 323 and 327 amino acids, respectively. As with all highly dynamic and specific regulatory proteins, PP1 plays an important role in cellular processes (such as the cell cycle, protein synthesis, and transcription), as well as pathologies (such as cancer, heart disease, memory loss, type 2 diabetes, and viral infections) in which it has potential therapeutic roles [15][16][17].
PIPs (PP1-interacting proteins) interact with the different PP1 regulatory subunits through short sequences, which create PP1 holoenzymes with unique properties, a strategy that has been defined as a molecular “lego” that governs specificity [10]. Initially, the RXvF-like motif was defined as a region of five amino acids with the consensus sequence of [K/R]-X(0,1)-[VI]-(P)-[FW], where X is any residue and P is any residue except proline [18]. However, the sequence is summarized as [KR]-[KR]-[VI]-(FIMYDP)-[FW], where the most conserved residues are 1, 3, and 5; at position 4, the following amino acids are never found: Phe (F), Ile (I), Met (M), Tyr (Y), Asp (D), and Pro (P) [9]. However, this sequence is mostly enriched for R (17%), K (11%), S (21%), and T (18%). The presence of S and T is decisive in the interaction between PP1 and PIPs because the phosphorylation of these residues disrupts the union with PP1 [9][19]. The docking of the RVxF domain does not affect the catalytic activity of PP1, but it does increase the concentration of the interactor, which alters the substrate specificity of PP1. On the other hand, crystallographic studies have shown that the RVxF domain of a PIP is found in a flexible loop that adopts an extended beta-sheet by binding to the hydrophobic pocket of PP1, which is 20 Å farther from the catalytic site [9][19]. According to the structure predicted in AlphaFold, IIIG9 exhibits the RKVHF sequence between amino acids 129 to 133 at the level of a short helix present in the human sequence. Furthermore, this sequence lacks the S/T amino acids at position 4, indicating that the interaction between IIIG9 (Figure 1) and the catalytic subunits of PP1 is not regulated by phosphorylation/dephosphorylation. Although some PP1 regulatory proteins specifically interact with a single PP1 isoform, others bind all the catalytic subunits of this phosphatase, at least in vitro [10]. Double hybrid assays performed with a cDNA library derived from human testes and using a bait for the union of the PP1γ1 and PP1γ2 isoforms (generated by tissue-specific alternative splicing of the PP1γ gene) demonstrated that human IIIG9 interacts with both isoforms [20]. In turn, the colocalization of IIIG9 and PP1γ has been observed in spermatogonia and mature spermatozoa from mice and bovines, respectively [21]. In addition, the interaction between IIIG9 and the alpha subunit of PP1 (PP1α) has been demonstrated via co-immunoprecipitation studies on HEK293 cell extracts that overexpress C11orf66-GFP (human IIIG9 fused to GFP) and the use of yeast co-transformation assays [9][20].
The identification of new regulatory subunits of the catalytic subunits of PP1 will allow researchers to understand how the functional diversity of this phosphatase is regulated in eukaryotic cells. In addition, the presence of numerous PIPs in concentrations that are in large molar excess prevents PP1 from being free, causing uncontrolled dephosphorylation that leads to cell death [22]. Additionally, the characterization of the expression and function of new PIPs, such as IIIG9, will make it possible to define the molecular interactome of PP1 around a certain cellular process, such as during the formation of cell polarity where PP1 participates in signaling mechanisms with Cdc42/Par-3/aPKC/Par6 [9], opening new therapeutic targets that specifically regulate the action of PP1. Other interaction motifs located near the N-terminus of the RKVHF sequence, such as MyPhoNE, RxxQ[VIL][KR]x[YW], SILK [GS]IL[RK], SpiDoC [10], and IDoHA [10], have been characterized in other PIPs, but these are not present in IIIG9 [9].
4. Expression of IIIG9 during CNS Development
The early and high expression of IIIG9 in the ventricular and cortical wall at embryonic day 17 of the rat brain has been demonstrated by immunocytochemical studies in rat brains [23]. At this stage, IIIG9 is widely expressed throughout the ventricular and cortical thickness, and it is preferentially polarized at the apical border of the lateral ventricle, which houses the body of the neural stem cell, called the radial glia. Researchers' localization results are also supported by data present in the CORTECON information base, which is a repository of cortical developmental gene expression, showing that the expression of IIIG9 mRNA (PPP1R32) is associated with neural differentiation states and the cortical specification of the upper cortical layers [24][25]. Moreover, evidence suggests that IIIG9 could participate in the development of neural stem cells (NSCs) by maintaining the signaling pathways for cell survival and differentiation, as demonstrated by DICER (riboendonuclease in the small RNA pathway) loss-of-function studies in NSCs, which increases anti-survival and/or apoptotic proteins and signaling pathways, thus leading to NSC cell death in the absence of mitogens [26]. Proteomic analysis showed that the levels of approximately 2900 proteins, including IIIG9, are affected. Therefore, the proteins lost in DICER-null NSCs could be relevant to or a consequence of the deregulation of cell survival and differentiation signaling pathways in NSCs or radial glia. Similarly, the location of apically polarized IIIG9 in the radial glia of the embryonic ventricular wall may be indicative of its role in cell polarization mechanisms that maintain adherens junctions or induce the multiciliogenesis of the radial glia that become ependymal cells. The general view is that multiciliated cells arise from progenitors after the inhibition of Notch, a process that triggers the activation of the master regulators of multiciliogenesis, GEMC1, and multicilin, which activate gene expression by binding the transcription factors, E2F4/5 and DP1 [27][28]. Additionally, the downstream binding of the transcription factor RFX2/3 mediates the biogenesis of both primary and motile cilia, while the transcription factor FOXJ1 anchors basal bodies, axoneme growth, and ciliary motility [29][30][31]. Notably, the analysis of the IIIG9 gene locus in mice (chromosome 19) using the GTRD [32][33], IFTI [34], and PROMO [35] databases has identified the binding sites for the transcription factors RFX2/3 and FOXJ1 immediately and 408 bp upstream of the start codon, respectively. This in silico analysis complements expression and localization data, strengthening the idea that IIIG9 is involved in the multiciliated program. Furthermore, a ~14-fold increase in IIIG9 mRNA levels was detected just 5 h after the induction of multicilin in epithelial explants of Xenopus laevis, positioning IIIG9 as one of the top 15 genes upregulated among >500 mRNAs after treatment [27].
IIIG9 could also be part of the basal body of the primary cilium of the radial glia that contacts the ventricular cavity, given that the IIIG9–GFP fusion protein was shown to be localized at the base of the cilium from the mouse kidney collecting duct cell line, IMCD3 [36]. This suggests that IIIG9 is not an exclusive protein of motile cilia, as it can also be present in the primary cilium of different cells of an organism. Thus, like many ciliary proteins, IIIG9 has extraciliary functions, such as cell cycle regulation in tumor cells, the maintenance of adherens junctions in ependymal epithelium, and the genesis and differentiation of neuronal subpopulations during the development of the cerebral cortex.
5. Role of IIIG9 in Pathologies
Although there is little information that directly relates the deregulation of the C11orf66 gene or the IIIG9 protein with the development of human pathologies, the following sections summarize what has been reported to date.
5.1. Ciliopathies
Ciliopathies are a set of developmental (degenerative) disorders of a single gene whose dysfunction alters the function of any type of cilia. According to the “Rare Diseases AutoRIF ARCHS4 Predictions” database [37][38], PPP1R32 dysfunction is postulated to be connected with the development of bronchiectasis-ciliary dyskinesia, cranioectodermal dysplasia, multifocal heterotopia, primary ciliary dyskinesia, situs inversus, and ciliary immobility associated with sterility. Similarly, the ciliary localization of IIIG9 has been demonstrated in human airway epithelial cells [39]. Furthermore, the C11orf66 gene is one of the differentially expressed genes in the pseudoglandular-to-canalicular transition of human lung development [40], which could have broader implications in terms of lung function in both health and illness. Similarly, researchers have shown that the loss of IIIG9 in the adult ventricular wall of the nervous system induces the presence of non-polarized ependymal cells (balloon-like morphology) and ependymal cells with rigid cilia that probably vibrate, which may be the basis of the observed ventriculomegaly [41], and are key agents in the development of a ciliopathy, such as hydrocephalus.
5.2. Infertility
IIIG9 was first reported as a ciliary protein in the testes and oviducts [23][39]. In the testes, IIIG9 has been detected in mouse spermatogonia (GC-1), uniformly in the nucleus and cytoplasm; in mature bovine spermatozoa, IIIG9 is absent in the equatorial region of the head and midpiece, standing out in the tail and head and constituting a regulatory protein of interest in sperm development [21]. To date, the effects of IIIG9 inhibition on sperm development and capacitation are unknown. Both processes were studied using an ex vivo model of rat seminiferous tubules against exposure to two fungicides (carbendazim and iprodione) that produce a strong alteration of spermatogenesis that affects the migration of germ cells through the seminiferous tubules; it also impacts androgen synthesis and alters meiosis. In this entry, increased IIIG9 expression was observed after 14 days of exposure to carbendazim (50 nM), decreasing after 21 days of exposure when combined with iprodione in equimolar proportions. In addition, IIIG9 was part of a series of proteins that were identified as potential markers of testicular dysfunction and infertility [42].
The targeted interruption of the Ppp1cc gene (PP1 gamma) also causes infertility in mice as a result of altered spermatogenesis coinciding with a loss of the transition between round and elongated spermatogonia and a decrease in germ cells, especially spermatids. These effects culminate in the generalized absence of sperm, resulting from apoptosis that occurs in all layers of the seminiferous tubules [43][44][45][46]. Thus, IIIG9 is likely a PP1 gamma regulatory protein that regulates its function to promote normal spermatogenesis.
5.3. Autism Spectrum Disorder (ASD)
ASD is a condition related to brain development that affects how the person perceives and socializes with other people, leading to problems in social interaction and communication, as well as the manifestation of restricted or repetitive behaviors, interests, and activities. Mak et al., 2017 carried out chromosomal microarray analysis in a cohort of 258 Chinese children with ASD, analyzing the copy number variants (CNVs) to identify possible molecular markers that predispose ASD development [47]. This entry was focused on nine patients with pathogenic and probably pathogenic CNVs who manifested ASD at different ages. In one patient (number 9), who was diagnosed at 29 months of age without physical abnormalities and with normal cognitive function, there were two consecutive duplications in a mosaic pattern, 11q12.1-q12. 2 (5.55 Mb) and 11p11.2 (0.29 Mb), which extended through the centromere [47]. In the first duplication (5.55 Mb region), the PPP1R32 (IIIG9) gene was one of the 64 genes present in this CNV that was classified as pathogenic and with unknown inheritance. Furthermore, this result was indicative of a marker chromosome, confirmed by karyotyping mos 47,XY,+mar/46,XY. Another patient with similar duplications (variant ID NSSV582454 present in the Children’s Hospital of Philadelphia database) presented a global developmental delay (GDD) and a delay in speech and language development [47]. These results could be related to the putative role of IIIG9 in the neural precursors of the cerebral cortex due to the high expression of this gene observed in the CORTECON database. In this way, questions arise regarding to the functional importance of IIIG9 during brain development and how the deregulation of this protein can contribute to the development of pathologies and developmental disorders, such as ASD.
5.4. Drug Use Disorders
Two reports have related IIIG9 expression to drug use disorders, such as alcoholism and cocaine use. Alcoholism is a complex disease consisting of the inability to control alcohol consumption due to physical and mental dependence. Although it has been established that genetic and environmental factors are crucial for its manifestation, the interaction between them is currently unknown. The screening analysis of the single-nucleotide polymorphisms (SNPs) of alcoholism using an Ensemble Bayesian Network (an important method used to analyze SNPs related to complex diseases) was used to identify the IIIG9 gene (C11orf66) among 18 genes that are potentially correlated with vulnerability to alcoholism [48]. Interestingly, the authors of this entry also identified compounds that could be used as potential medicines for the treatment of alcoholism since they regulate the expression of at least 14 of the 18 initially recognized genes. For C11orf66, 13 compounds were identified: four strongly increase gene expression, and five, including chloroprene, inhibit expression [48]. However, the carcinogenic and mutagenic effects of chloroprene limit its use in future molecular strategies to regulate the expression of IIIG9 in patients with alcoholism.
The mesolimbic system also plays an important role in neuroadaptation against drug use, mainly in the reward circuit that connects the ventral tegmental area (VTA) and the nucleus accumbens (NAc) [49]. In an RNA-seq transcriptomic analysis study, changes in VTA and NAc expression were analyzed in Rhesus macaques exposed to cocaine self-administration (3 months at a maximum of 3 mg/kg weight) and compared to a control group. IIIG9 (PPP1R32) was differentially expressed (q < 0.05) in the VTA of animals that consumed cocaine [50]. This increase in the expression of some genes is attributed to epigenetic changes and chromatin remodeling at the level of the dopaminergic system, and it is related to the transition from drug use to abuse [51]. In this way, IIIG9 is considered a gene of interest in understanding the molecular and cellular mechanisms of addiction. Future studies will help to understand whether regulating IIIG9 function can become a potential treatment strategy.
References
- Danielson, P.E.; Sautkulis, L.N.; Foye, P.E.; Hedlund, P.B.; Carson, M.J. A novel mRNA expressed along brain ventricles. Brain Res. Gene Expr. Patterns 2002, 1, 83–88.
- Nucleotide . Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information . Accession No. NM_145017.3, NM_001170753.2, Homo Sapiens Protein Phosphatase 1 Regulatory Subunit 32 (PPP1R32), mRNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NM_145017.3,NM_001170753.2 (accessed on 10 June 2022).
- Nucleotide . Bethesda (MD): National Library of Medicine (US), National Center for Biotechnology Information . Accession No. NM_133689.2, NM_001368184.1, XM_006527302.5, XM_017318281.1, XM_017318282.1, XM_036161672.1, Mus Musculus Protein Phosphatase 1 Regulatory Subunit 32 (Ppp1r32), mRNA. Available online: https://www.ncbi.nlm.nih.gov/nuccore/NM_133689.2,NM_001368184.1,XM_006527302.5,XM_017318281.1,XM_017318282.1,XM_036161672.1 (accessed on 10 June 2022).
- C-I-TASSER (Contact-guided Iterative Threading ASSEmbly Refinement) Server. Available online: https://zhanggroup.org/C-I-TASSER/ (accessed on 10 June 2022).
- Zheng, W.; Zhang, C.; Li, Y.; Pearce, R.; Bell, E.W.; Zhang, Y. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep. Methods 2021, 1, 100014.
- AlphaFold Protein Structure Database, Developed by DeepMind and EMBL-EBI. Available online: https://alphafold.ebi.ac.uk/(accessed on 10 June 2022).
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589.
- Varadi, M.; Anyango, S.; Deshpande, M.; Nair, S.; Natassia, C.; Yordanova, G.; Yuan, D.; Stroe, O.; Wood, G.; Laydon, A.; et al. AlphaFold Protein Structure Database: Massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 2022, 50, D439–D444.
- Hendrickx, A.; Beullens, M.; Ceulemans, H.; Den Abt, T.; Van Eynde, A.; Nicolaescu, E.; Lesage, B.; Bollen, M. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 2009, 16, 365–371.
- Heroes, E.; Lesage, B.; Gornemann, J.; Beullens, M.; Van Meervelt, L.; Bollen, M. The PP1 binding code: A molecular-lego strategy that governs specificity. FEBS J. 2013, 280, 584–595.
- Reynolds, C.R.; Islam, S.A.; Sternberg, M.J.E. EzMol: A Web Server Wizard for the Rapid Visualization and Image Production of Protein and Nucleic Acid Structures. J. Mol. Biol. 2018, 430, 2244–2248.
- EzMol Interface © Structural Bioinformatics Group, Imperial College London 2019. Available online: http://www.sbg.bio.ic.ac.uk/~ezmol/ (accessed on 10 June 2022).
- Casamayor, A.; Arino, J. Controlling Ser/Thr protein phosphatase PP1 activity and function through interaction with regulatory subunits. Adv. Protein Chem. Struct. Biol. 2020, 122, 231–288.
- Verbinnen, I.; Ferreira, M.; Bollen, M. Biogenesis and activity regulation of protein phosphatase 1. Biochem. Soc. Trans. 2017, 45, 89–99.
- Figueiredo, J.; da Cruz, E.S.O.A.; Fardilha, M. Protein phosphatase 1 and its complexes in carcinogenesis. Curr. Cancer Drug Targets 2014, 14, 2–29.
- Felgueiras, J.; Jeronimo, C.; Fardilha, M. Protein phosphatase 1 in tumorigenesis: Is it worth a closer look? Biochim. Biophys. Acta Rev. Cancer 2020, 1874, 188433.
- Ferreira, M.; Beullens, M.; Bollen, M.; Van Eynde, A. Functions and therapeutic potential of protein phosphatase 1: Insights from mouse genetics. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 16–30.
- Ceulemans, H.; Bollen, M. A tighter RVxF motif makes a finer Sift. Chem. Biol. 2006, 13, 6–8.
- Egloff, M.P.; Johnson, D.F.; Moorhead, G.; Cohen, P.T.; Cohen, P.; Barford, D. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 1997, 16, 1876–1887.
- Fardilha, M.; Esteves, S.L.; Korrodi-Gregorio, L.; Vintem, A.P.; Domingues, S.C.; Rebelo, S.; Morrice, N.; Cohen, P.T.; da Cruz e Silva, O.A.; da Cruz e Silva, E.F. Identification of the human testis protein phosphatase 1 interactome. Biochem. Pharm. 2011, 82, 1403–1415.
- Sousa, C.; Vintém, A.P.B.; Fardilha, M.; Silva, O.d.C.e.; Silva, E.F.d. Subcellular Localization of a Novel Alternative Splicing of IIIG9 and Colocalization with PPP1gamma Isoforms. Microsc. Microanal. 2008, 14, 141–143.
- Chatterjee, J.; Beullens, M.; Sukackaite, R.; Qian, J.; Lesage, B.; Hart, D.J.; Bollen, M.; Kohn, M. Development of a peptide that selectively activates protein phosphatase-1 in living cells. Angew. Chem. Int. Ed. Engl. 2012, 51, 10054–10059.
- Cifuentes, M.; Baeza, V.; Arrabal, P.M.; Visser, R.; Grondona, J.M.; Saldivia, N.; Martinez, F.; Nualart, F.; Salazar, K. Expression of a Novel Ciliary Protein, IIIG9, During the Differentiation and Maturation of Ependymal Cells. Mol. Neurobiol. 2018, 55, 1652–1664.
- van de Leemput, J.; Boles, N.C.; Kiehl, T.R.; Corneo, B.; Lederman, P.; Menon, V.; Lee, C.; Martinez, R.A.; Levi, B.P.; Thompson, C.L.; et al. CORTECON: A temporal transcriptome analysis of in vitro human cerebral cortex development from human embryonic stem cells. Neuron 2014, 83, 51–68.
- CORTECON. Repository of Gene Expression in the Developing Cortex. NSCI, Neural Stem Cell Institute. Gene Expression: PPP1R32. Available online: https://cortecon.neuralsci.org/index.php?cort_mode=genedisplay&geneid=220004 (accessed on 10 June 2022).
- Kawase-Koga, Y.; Low, R.; Otaegi, G.; Pollock, A.; Deng, H.; Eisenhaber, F.; Maurer-Stroh, S.; Sun, T. RNAase-III enzyme Dicer maintains signaling pathways for differentiation and survival in mouse cortical neural stem cells. J. Cell Sci. 2010, 123, 586–594.
- Stubbs, J.L.; Vladar, E.K.; Axelrod, J.D.; Kintner, C. Multicilin promotes centriole assembly and ciliogenesis during multiciliate cell differentiation. Nat. Cell Biol. 2012, 14, 140–147.
- Terre, B.; Piergiovanni, G.; Segura-Bayona, S.; Gil-Gomez, G.; Youssef, S.A.; Attolini, C.S.; Wilsch-Brauninger, M.; Jung, C.; Rojas, A.M.; Marjanovic, M.; et al. GEMC1 is a critical regulator of multiciliated cell differentiation. EMBO J. 2016, 35, 942–960.
- Choksi, S.P.; Lauter, G.; Swoboda, P.; Roy, S. Switching on cilia: Transcriptional networks regulating ciliogenesis. Development 2014, 141, 1427–1441.
- Choksi, S.P.; Babu, D.; Lau, D.; Yu, X.; Roy, S. Systematic discovery of novel ciliary genes through functional genomics in the zebrafish. Development 2014, 141, 3410–3419.
- Meunier, A.; Azimzadeh, J. Multiciliated Cells in Animals. Cold Spring Harb. Perspect. Biol. 2016, 8, a028233.
- Yevshin, I.; Sharipov, R.; Valeev, T.; Kel, A.; Kolpakov, F. GTRD: A database of transcription factor binding sites identified by ChIP-seq experiments. Nucleic Acids Res. 2017, 45, D61–D67.
- Kolmykov, S.; Yevshin, I.; Kulyashov, M.; Sharipov, R.; Kondrakhin, Y.; Makeev, V.J.; Kulakovskiy, I.V.; Kel, A.; Kolpakov, F. GTRD: An integrated view of transcription regulation. Nucleic Acids Res. 2021, 49, D104–D111.
- Ghosh, D. Object-oriented transcription factors database (ooTFD). Nucleic Acids Res. 2000, 28, 308–310.
- Farre, D.; Roset, R.; Huerta, M.; Adsuara, J.E.; Rosello, L.; Alba, M.M.; Messeguer, X. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. 2003, 31, 3651–3653.
- Sigg, M.A.; Menchen, T.; Lee, C.; Johnson, J.; Jungnickel, M.K.; Choksi, S.P.; Garcia, G., 3rd; Busengdal, H.; Dougherty, G.W.; Pennekamp, P.; et al. Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways. Dev. Cell 2017, 43, 744–762.
- Lachmann, A.; Torre, D.; Keenan, A.B.; Jagodnik, K.M.; Lee, H.J.; Wang, L.; Silverstein, M.C.; Ma’ayan, A. Massive mining of publicly available RNA-seq data from human and mouse. Nat. Commun. 2018, 9, 1366.
- ARCHS4: Massive Mining of Publicly Available RNA-seq Data from Human and Mouse. Gene PPP1R32. Available online: https://maayanlab.cloud/archs4/gene/PPP1R32 (accessed on 10 June 2022).
- Ivliev, A.E.; t Hoen, P.A.; van Roon-Mom, W.M.; Peters, D.J.; Sergeeva, M.G. Exploring the transcriptome of ciliated cells using in silico dissection of human tissues. PLoS ONE 2012, 7, e35618.
- Sharma, S.; Tantisira, K.; Carey, V.; Murphy, A.J.; Lasky-Su, J.; Celedon, J.C.; Lazarus, R.; Klanderman, B.; Rogers, A.; Soto-Quiros, M.; et al. A role for Wnt signaling genes in the pathogenesis of impaired lung function in asthma. Am. J. Respir. Crit. Care Med. 2010, 181, 328–336.
- Baeza, V.; Cifuentes, M.; Martinez, F.; Ramirez, E.; Nualart, F.; Ferrada, L.; Oviedo, M.J.; De Lima, I.; Troncoso, N.; Saldivia, N.; et al. IIIG9 inhibition in adult ependymal cells changes adherens junctions structure and induces cellular detachment. Sci. Rep. 2021, 11, 18537.
- Pisani, C.; Voisin, S.; Arafah, K.; Durand, P.; Perrard, M.-H.; Guichaoua, M.-R.; Bulet, P.; Prat, O. Ex vivo assessment of testicular toxicity induced by carbendazim and iprodione, alone or in a mixture. ALTEX Altern. Anim. Exp. 2016, 33, 393–413.
- Varmuza, S.; Jurisicova, A.; Okano, K.; Hudson, J.; Boekelheide, K.; Shipp, E.B. Spermiogenesis is impaired in mice bearing a targeted mutation in the protein phosphatase 1cgamma gene. Dev. Biol. 1999, 205, 98–110.
- Oppedisano-Wells, L.; Varmuza, S. Protein phosphatase 1cgamma is required in germ cells in murine testis. Mol. Reprod. Dev. 2003, 65, 157–166.
- Chakrabarti, R.; Kline, D.; Lu, J.; Orth, J.; Pilder, S.; Vijayaraghavan, S. Analysis of Ppp1cc-null mice suggests a role for PP1gamma2 in sperm morphogenesis. Biol. Reprod. 2007, 76, 992–1001.
- Chakrabarti, R.; Cheng, L.; Puri, P.; Soler, D.; Vijayaraghavan, S. Protein phosphatase PP1 gamma 2 in sperm morphogenesis and epididymal initiation of sperm motility. Asian J. 2007, 9, 445–452.
- Mak, A.S.L.; Chiu, A.T.G.; Leung, G.K.C.; Mak, C.C.Y.; Chu, Y.W.Y.; Mok, G.T.K.; Tang, W.F.; Chan, K.Y.K.; Tang, M.H.Y.; Lau Yim, E.T.; et al. Use of clinical chromosomal microarray in Chinese patients with autism spectrum disorder-implications of a copy number variation involving DPP10. Mol. Autism 2017, 8, 31.
- Chen, H.; Sun, J.; Jiang, H.; Wang, X.; Wu, L.; Wu, W.; Wang, Q. Inferring Alcoholism SNPs and Regulatory Chemical Compounds Based on Ensemble Bayesian Network. Comb. Chem. High. Throughput Screen 2017, 20, 107–115.
- Nestler, E.J. Is there a common molecular pathway for addiction? Nat. Neurosci. 2005, 8, 1445–1449.
- Vallender, E.J.; Goswami, D.B.; Shinday, N.M.; Westmoreland, S.V.; Yao, W.D.; Rowlett, J.K. Transcriptomic profiling of the ventral tegmental area and nucleus accumbens in rhesus macaques following long-term cocaine self-administration. Drug Alcohol Depend. 2017, 175, 9–23.
- Schmidt, H.D.; McGinty, J.F.; West, A.E.; Sadri-Vakili, G. Epigenetics and psychostimulant addiction. Cold Spring Harb. Perspect. Med. 2013, 3, a012047.
More
Information
Subjects:
Cell Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
888
Revisions:
2 times
(View History)
Update Date:
01 Dec 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




