Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Zeinab Ezzeddine | -- | 1889 | 2022-11-29 06:48:19 | | | |
| 2 | Dean Liu | Meta information modification | 1889 | 2022-11-30 02:53:42 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Ghssein, G.; Ezzeddine, Z. Pseudomonas aeruginosa Metallophores. Encyclopedia. Available online: https://encyclopedia.pub/entry/37005 (accessed on 19 January 2026).
Ghssein G, Ezzeddine Z. Pseudomonas aeruginosa Metallophores. Encyclopedia. Available at: https://encyclopedia.pub/entry/37005. Accessed January 19, 2026.
Ghssein, Ghassan, Zeinab Ezzeddine. "Pseudomonas aeruginosa Metallophores" Encyclopedia, https://encyclopedia.pub/entry/37005 (accessed January 19, 2026).
Ghssein, G., & Ezzeddine, Z. (2022, November 29). Pseudomonas aeruginosa Metallophores. In Encyclopedia. https://encyclopedia.pub/entry/37005
Ghssein, Ghassan and Zeinab Ezzeddine. "Pseudomonas aeruginosa Metallophores." Encyclopedia. Web. 29 November, 2022.
Copy Citation
The human pathogen Pseudomonas aeruginosa (P. aeruginosa) causes several infections, both acute and chronic, mainly in hosts with compromised immunity and in patients suffering from cystic fibrosis. The pathogenesis of this bacterium is caused by several factors.
Pseudomonas aeruginosa
metallophores
pathogenesis
1. Introduction
The Pseudomonas genus comprises more than 120 species of rod shaped, Gram-negative and flagellated bacteria that are predominant in humid environments including water and soil [1][2]. These bacteria can be pathogenic to humans, plants and animals [3]. Gram-negative bacteria have two membranes, a peptidoglycan thin layer encircled by an outer membrane containing lipopolysaccharide (LPS) [4]. The latter is made up of three units: a hydrophilic polysaccharide, O antigen, and lipid A, which is a hydrophobic domain, responsible for the endotoxic activity of these pathogens [5]. Pseudomonas aeruginosa (P. aeruginosa) strains mostly associated with causing human infection are γ-Proteobacterium [6]. P. aeruginosa causes serious nosocomial infections, in addition to infections that can be fatal in immunocompromised people and persons suffering from physical damages (e.g., burn wounds) in addition to chronic infections in patients with cystic fibrosis [7][8]. In addition, it causes many other diseases ranging from chronic respiratory tract to urinary tract infections and blood infections [9]. This opportunistic bacterium was considered a significant threat by the Centers for Disease Control and Prevention (CDC) in a 2013 antibiotic resistance threat report [10] as a result of the increasing development of multidrug resistant (MDR) strains leading to a lot of therapeutic challenges due to the absence of effective treatment and consequently high rates of mortality [11][12]. The outer membrane permeability of P. aeruginosa is low, acting as an intrinsic barrier and making it resistant to several antibiotics [13]. When subjected to antibiotic pressure, this bacterium is able to adapt to this condition through an enhanced metabolic response that promotes the bacterial survival and antibiotic resistance development [14]. Because this bacterium has low nutritional requirements and adaptable energy metabolism, it can adjust to conditions that other organisms cannot tolerate [15]. Moreover, strains of P. aeruginosa can form a biofilm making their cells insensitive to disinfectants or host defense mechanisms [16]. Such pathogenesis results from the production of various virulence factors during infection, permitting it to survive and colonize its hosts [17]. Some of these factors are: type IV pili (the main adhesion to epithelial cells) [18], Exotoxin A (toxin that causes host cells death by necrosis) [19] and metallophores (secondary metabolites for metal ions sequestering) [20]. Like other bacteria, P. aeruginosa directly interacts with the extracellular medium in order to control and constantly maintain the intracellular metal ions concentration [21]. Metals are involved in their development and survival through a regulated network that includes several specific metal–DNA binding transcriptional regulators [22]. Most enzymes utilize metal ions as co-factors, and when the availability of one essential metal ion is low, some essential enzymes needed for metabolism cannot function properly, thus affecting cellular growth. On the other hand, excess metal ions can cause toxicity even if they are necessary for bacterial survival [23].
P. aeruginosa has powerful metal acquisition pathways enabling it to withstand growth conditions changes. They produce metallophores that chelate metal ions from the surrounding environment especially in minimum media. One of the strategies used by P. aeruginosa to uptake iron, which is a key element for this bacterium pathogenesis, is siderophores production. The latter are small ferric iron ions chelators that can scavenge and transport iron ions from the extracellular environment via specific protein receptors located on the outer membrane [24][25].
The first siderophore is pyoverdine; it has high iron chelating affinity, and it has been reported that strains unable to produce pyoverdines had reduced virulence during mice infections [26]. The importance of pyoverdines in the virulence of this bacterium was also proved using mouse and rabbit lung infection models [27][28]. It was found that pyoverdines play a dual role during infection. In addition to iron scavenging, they act as signaling molecules for the production of two vital virulence factors (the endo-proteinase PrpL and exotoxin A) [29][30]. The other siderophore produced by this bacterium is pyochelin, although its iron affinity is lower than that of pyoverdine [31] and fewer genes are involved in its biosynthesis [32]. It has been indicated that pyochelin is firstly produced by P. aeruginosa, but only when iron concentration becomes extremely low does this bacterium starts producing pyoverdine [33]. Pyochelin production might have a role in the continuous inflammatory response that cause tissues damage in chronic infections, such as in cystic fibrosis lungs [34]. P. aeruginosa also produces a third metallophore specific for the uptake of metal ions other than iron. It is a narrow spectrum metallophore called pseudopaline [35]. Pseudopaline is related to the nicotianamine of plants and it is a an opine-type metallophore that belongs to the same family of staphylopine synthetized by Staphylococcus aureus (S. aureus) [36].
2. Pyoverdine
Iron is crucial for bacterial cellular function, but at physiological pH (near neutral), iron has low solubility and is not available freely to pathogens. This low bioavailability leads to competitive interactions for iron between host and pathogen [37]. Pathogens have evolved sensitive systems for detecting low intracellular iron and stimulating siderophore production [38][39].
Pyoverdines were first discovered in 1892 [40], and their role in the acquisition of iron by Pseudomonads was indicated in late 1970s [41]. Currently, over 100 pyoverdines secreted by different Pseudomonas strains and species have been identified [42] which represent about 20% of the characterized microbial siderophores [43]. Their general chemical structure contains three bidentate chelating sites, including two hydroxamates and a catechol (Figure 1). These siderophores are made up of three parts: a peptide specific for each strain with a sequence of 6 to 12 amino acids linked to the carboxyl group, a chromophore part derived from 2,3-diamino-6,7-dihydroxyquinoline which is responsible for their fluorescence [44], and a side-chain linked at C-3 position to the chromophore nitrogen atom (NH2 group). In the majority of cases, the side-chain is a Krebs cycle diacid, such as malic, succinic or one of their amide derivatives [45].
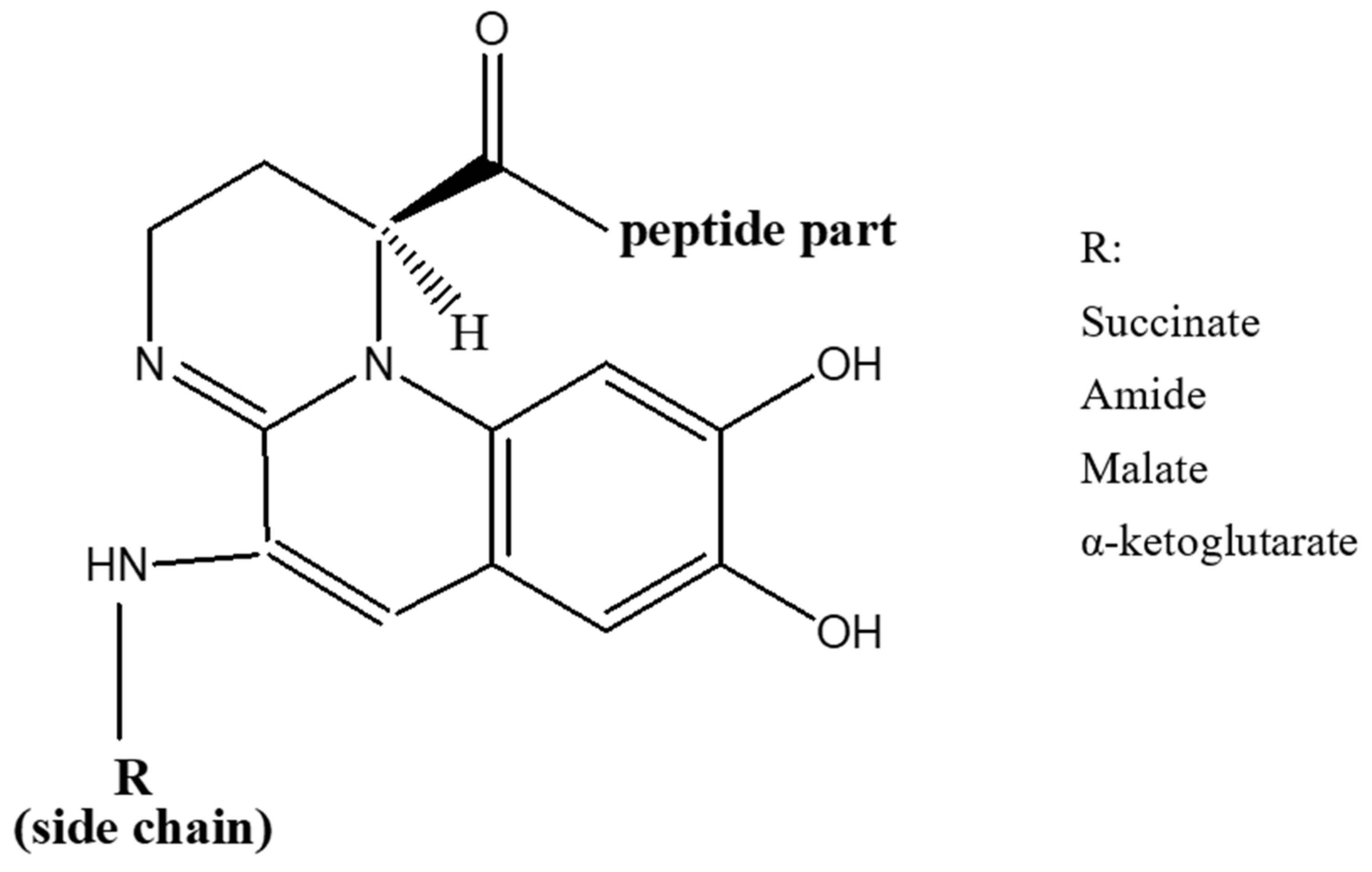
Figure 1. General structure of pyoverdines.
The conserved chromophore part among all Pseudomonas species and strains chelates Fe3+ via the function catechol. The peptide moiety interacts with Fe3+ via two functions (hydroxamate and/or hydroxycarboxylate) and its sequence differs between each Pseudomonads species as well as strains of the same species [46]. Moreover, it can be linear or cyclic, and comprise unusual amino acids (e.g., D-isomers) and others that are not usually found in biomolecules [47].
Three structurally distinct types of pyoverdine are produced by P. aeruginosa strains (class I, II and III); each has its characteristic peptide chain [47]. The Pyoverdines produced by fluorescent P. aeruginosa strains are presented in Table 1.
Table 1. Pyoverdines that are synthesized by P. aeruginosa strains.
| Species | Strains | Pyoverdines Type | Peptide Sequences | Ref |
|---|---|---|---|---|
| P. aeruginosa | ATCC15692(PAO1) | PVDI | Ser–Arg–Ser–FoOHOrn–[Lys–FoOHOrn–Thr–Thr] | [48] |
| P. aeruginosa | ATCC27853 | PVDII | Ser–FoOHOrn–Orn–Gly–Thr–Ser–cOHOrn | [49] |
| P. aeruginosa | Pa6 (R) (sv. III-l) | PVDIII-1 | Ser–Dab–FoOHOrn–Gln–Gln–FoOHOrn–Gly | [50] |
| P. aeruginosa | R’ (sv. III-2) | PVDIII-2 | Ser–Dab–FoOHOrn–Gln–FoOHOrn–Gly | [51] |
Dab: 2,4-diaminobutyrate; Orn: ornithine; OHOrn: N-hydroxyornithine; FoHOrn: N-formyl-N-hydroxyornithine; [ ] are used to show cyclic peptides.
Pyoverdines of class I (PVDI), produced by P. aeruginosa PAO1, are characterized by a cycle formed by the last four amino acids of the peptide moiety. An amide bond is formed between the carbonyl group of the C-terminal amino acid and the ε-amino group of an in-chain Lysine or Ornithine residue. On the other hand, the peptide sequence of class II pyoverdines (PVDII), produced by P. aeruginosa ATCC 27853, is linear with the C-terminal amino acid being of the N-hydroxy (cyclo)Ornithine type. As for class III pyoverdines (PVDIII), produced by P. aeruginosa Pa6, they have a linear peptide sequence with an unmodified N-hydroxyOrn at the C-terminus [47]. Iron acquisition mechanisms have been investigated the most in P. aeruginosa PAO1. A fourth pyoverdine that belongs to the strain R’ was also isolated where it was classified within the same siderovar (sv.) like strain Pa6 (sv. III). However, due to the variation in the structure of pyoverdine between the two strains, it was proposed to classify R’ strain in a sub-group (sv. III-2) of P. aeruginosa sv. III, different from the Pa6 sub-group (sv. III-l) [51].
3. Pyochelin
The second siderophore that is produced by P. aeruginosa is pyochelin (PCH) with lower iron affinity than pyoverdine as previously mentioned. It is a member of phenols, a monocarboxylic acid and a member of thiazolidines produced by salicylic acid condensation with two cysteine molecules [52]. The pyochelin structure is shown in Figure 2.
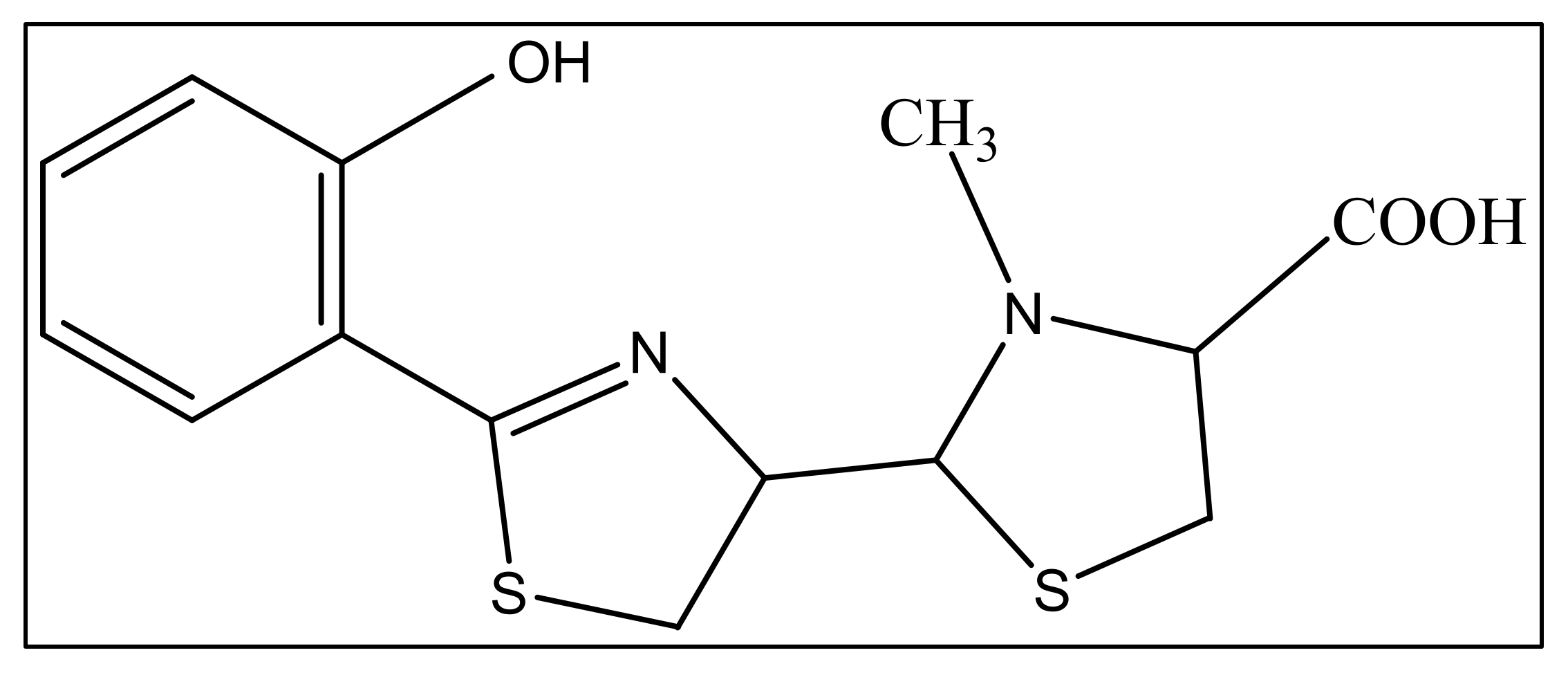
Figure 2. The structure of pyochelin.
4. Uptake of Siderophores Produced by Other Microorganisms
Besides pyoverdine (PVD) and pyochelin (PCH), P. aeruginosa detects the presence of exosiderophores excreted by other bacteria in its extracellular medium via sigma and anti-sigma factors which is an AraC family transcriptional regulator [53]. These regulators are responsible for the transcription activation of exosiderophore-corresponding TBDT (TonB-dependent Transporters) in the presence of Fe–exosiderophore as well as the proteins needed for iron release from the chelator once it enters the bacteria [54][55]. The exosiderophores presence induces both the transcription and expression of their corresponding TBDTs in iron limited medium. A study was done in order to investigate how P. aeruginosa acts in the presence of several exosiderophores such as Enterobactin (ENT), an E. coli catechol siderophore [56] and vibriobactin (VIB), which is another catechol siderophore secreted by Vibrio cholera [57]. These exosiderophores were found to be able to induce the transcription and expression of specific TBDTs and repressed the transcription and expression of fptA (PCH specific TBDT), along with all genes related to PCH pathway. On the other hand, no effect on the PVD pathway expression genes was observed, as well as the ferrous and heme uptake pathways, or those for any other TBDTs [58]. These results indicated that the bacterium utilizes the triscatechol siderophores to sequester iron, instead of their own siderophores. This great phenotypic plasticity related to diverse pathways for up taking iron present in the genome of P. aeruginosa, gives it a high adaptation potential of in a variety of biotopes [59].
5. Pseudopaline
In addition to iron, zinc is considered an essential metal ion required almost all living organisms at low concentrations [60], where it comes after iron in abundance [61]. In the opportunistic bacteria P. aeruginosa, it is well known that zinc plays an essential role in its virulence, host organism colonization, and antibiotic resistance [60]. Also, this bacteria needs zinc to resist carbapenems [62] as well as the extracellular proteases activity [63]. P. aeruginosa has several transport systems for zinc that enable it to survive in zinc scarce environments upon infection. In addition to PVD and pyochelin PCH, P. aeruginosa produces another metallophore called pseudopaline, biosynthesized by two enzymes, CntL and CntM [64]. Pseudopaline is plant nicotianamine related belonging to the same opine-type metallophore family as staphylopine and yersinopine synthetized by S. aureus and Y. pestis, respectively [36][65]. Various studies proved that pseudopaline contributes to the virulence of P. aeruginosa. The receptor CntA of pseudopaline is related to lung infections in cystic fibrosis patients, and the pseudopaline operon was highly upregulated in the infected host sites [66][67]. Besides zinc, pseudopaline is also involved in the uptake of cobalt [62] and nickel [35]. Urease, which is a nickel-dependent enzyme, is produced by P. aeruginosa [68]. Moreover, P. aeruginosa requires cobalt for the cobalamin-dependent ribonucleotide reductase (NrdJab) that is essential for developing biofilm under oxygen-limited conditions [69]. These findings demonstrate the importance of cobalt and nickel uptake in P. aeruginosa pathogenesis and the potential role of pseudopaline in sequestering these trace metal ions. The structure of pseudopaline is illustrated in Figure 3.
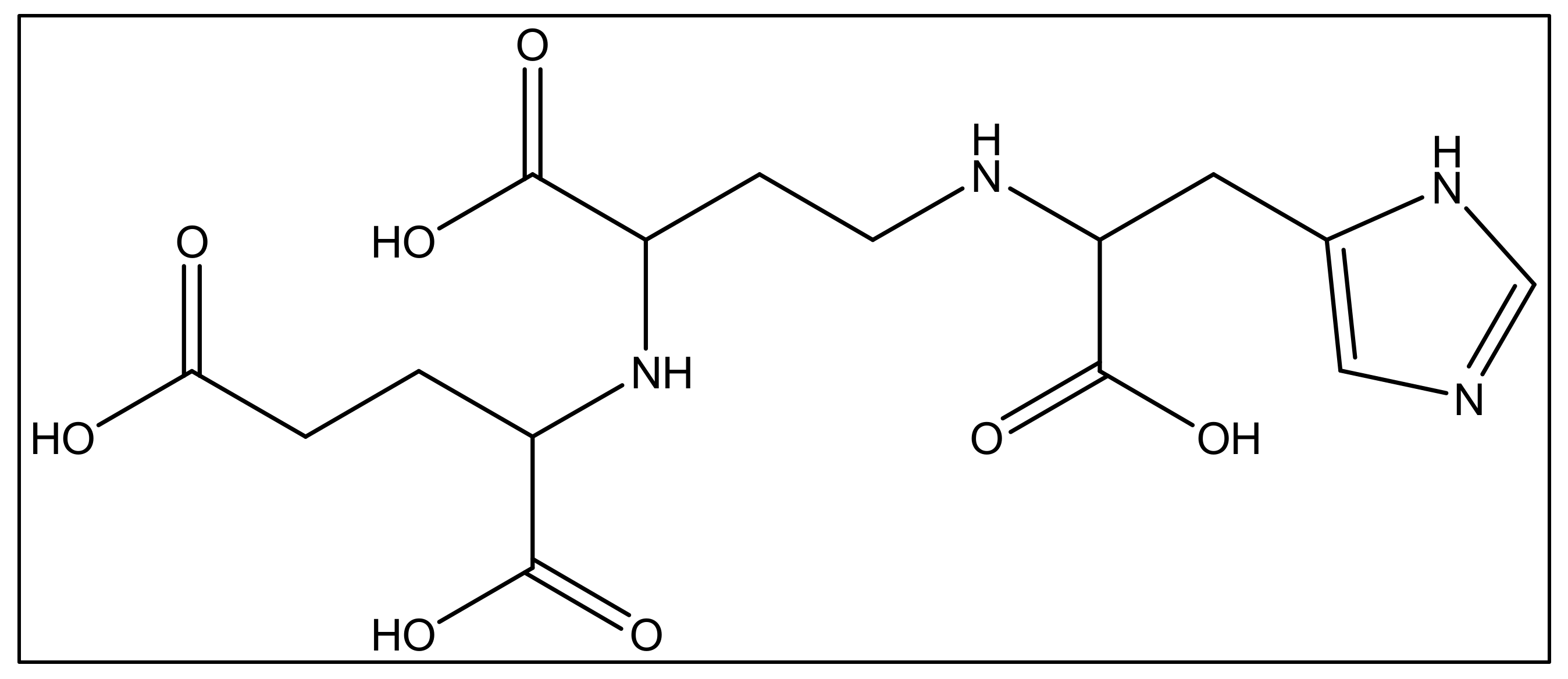
Figure 3. The structure of the opine-type metallophore in P. aeruginosa (pseudopaline).
References
- Peix, A.; Ramírez-Bahena, M.-H.; Velázquez, E. Historical evolution and current status of the taxonomy of genus Pseudomonas. Infect. Genet. Evol. 2009, 9, 1132–1147.
- Anzai, Y.; Kim, H.; Park, J.-Y.; Wakabayashi, H.; Oyaizu, H. Phylogenetic affiliation of the pseudomonads based on 16S rRNA sequence. Int. J. Syst. Evol. Microbiol. 2000, 50, 1563–1589.
- Wolfgang, M.C.; Kulasekara, B.R.; Liang, X.; Boyd, D.; Wu, K.; Yang, Q.; Miyada, C.G.; Lory, S. Conservation of genome content and virulence determinants among clinical and environmental isolates of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2003, 100, 8484–8489.
- Silhavy, T.J.; Kahne, D.; Walker, S. The Bacterial Cell Envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414.
- Livermore, D.M. Current Epidemiology and Growing Resistance of Gram-Negative Pathogens. Korean J. Intern. Med. 2012, 27, 128–142.
- Mahajan-Miklos, S.; Rahme, L.G.; Ausubel, F.M. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 2000, 37, 981–988.
- Winstanley, C.; O’Brien, S.; Brockhurst, M.A. Pseudomonas aeruginosa Evolutionary Adaptation and Diversification in Cystic Fibrosis Chronic Lung Infections. Trends Microbiol. 2016, 24, 327–337.
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa Lifestyle: A Paradigm for Adaptation, Survival, and Persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39.
- Neves, P.R.; McCulloch, J.A.; Mamizuka, E.M.; Lincopan, N. Aeruginosa. In Encyclopedia of Food Microbiology; Batt, C.A., Tortorello, M.-L., Eds.; Elsevier: Amsterdam, The Netherlands, 2014; Volume 3, pp. 253–260. ISBN 9780123847331.
- CDC. Antibiotic Resistance Threats in the United States; Centers for Disease Control and Prevention: Atlanta, GA, USA, 2013; 114p.
- Aloush, V.; Navon-Venezia, S.; Seigman-Igra, Y.; Cabili, S.; Carmeli, Y. Multidrug-Resistant Pseudomonas aeruginosa: Risk Factors and Clinical Impact. Antimicrob. Agents Chemother. 2006, 50, 43–48.
- Schechner, V.; Gottesman, T.; Schwartz, O.; Korem, M.; Maor, Y.; Rahav, G.; Karplus, R.; Lazarovitch, T.; Braun, E.; Finkelstein, R.; et al. Pseudomonas aeruginosa bacteremia upon hospital admission: Risk factors for mortality and influence of inadequate empirical antimicrobial therapy. Diagn. Microbiol. Infect. Dis. 2011, 71, 38–45.
- Breidenstein, E.B.; de la Fuente-Núñez, C.; Hancock, R.E. Pseudomonas aeruginosa: All roads lead to resistance. Trends Microbiol. 2011, 19, 419–426.
- Lambert, P.A. Mechanisms of antibiotic resistance in Pseudomonas aeruginosa. J. R. Soc. Med. 2002, 95, 22–26.
- Wu, M.; Guina, T.; Brittnacher, M.; Nguyen, H.; Eng, J.; Miller, S.I. The Pseudomonas aeruginosa Proteome during Anaerobic Growth. J. Bacteriol. 2005, 187, 8185–8190.
- Hall-Stoodley, L.; Costerton, J.W.; Stoodley, P. Bacterial biofilms: From the Natural environment to infectious diseases. Nat. Rev. Genet. 2004, 2, 95–108.
- Lazdunski, A. Regulation of virulence factors in Pseudomonas aeruginosa. Med. Mal. Infect. 1998, 28, 109–118.
- Hahn, H.P. The type-4 pilus is the major virulence-associated adhesin of Pseudomonas aeruginosa—A review. Gene 1997, 192, 99–108.
- Wick, M.; Frank, D.; Storey, D.; Iglewski, B. Structure, Function, and Regulation of Pseudomonas aeruginosa Exotoxin A. Annu. Rev. Microbiol. 1990, 44, 335–363.
- Kipnis, E.; Sawa, T.; Wiener-Kronish, J. Targeting mechanisms of Pseudomonas aeruginosa pathogenesis. Med. Mal. Infect. 2006, 36, 78–91.
- Guillon, L.; El Mecherki, M.; Altenburger, S.; Graumann, P.L.; Schalk, I.J. High cellular organization of pyoverdine biosynthesis in Pseudomonas aeruginosa: Clustering of PvdA at the old cell pole. Environ. Microbiol. 2012, 14, 1982–1994.
- Guerra, A.J.; Giedroc, D.P. Metal site occupancy and allosteric switching in bacterial metal sensor proteins. Arch. Biochem. Biophys. 2012, 519, 210–222.
- Foster, A.W.; Osman, D.; Robinson, N.J. Metal Preferences and Metallation. J. Biol. Chem. 2014, 289, 28095–28103.
- Cornelis, P. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 2010, 86, 1637–1645.
- Hider, R.C.; Kong, X. Chemistry and biology of siderophores. Nat. Prod. Rep. 2010, 27, 637–657.
- Meyer, J.M.; Neely, A.; Stintzi, A.; Georges, C.; Holder, A.I. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 1996, 64, 518–523.
- Xiong, Y.; Vasil, M.L.; Johnson, Z.; Ochsner, U.A.; Bayer, A.S. The Oxygen-and Iron-Dependent Sigma Factor pvdS of Pseudomonas aeruginosa Is an Important Virulence Factor in Experimental Infective Endocarditis. J. Infect. Dis. 2000, 181, 1020–1026.
- Minandri, F.; Imperi, F.; Frangipani, E.; Bonchi, C.; Visaggio, D.; Facchini, M.; Pasquali, P.; Bragonzi, A.; Visca, P. Role of Iron Uptake Systems in Pseudomonas aeruginosa Virulence and Airway Infection. Infect. Immun. 2016, 84, 2324–2335.
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077.
- Wilderman, P.J.; Vasil, A.I.; Johnson, Z.; Wilson, M.J.; Cunliffe, H.E.; Lamont, I.L.; Vasil, M.L. Characterization of an Endoprotease (PrpL) Encoded by a PvdS-Regulated Gene in Pseudomonas aeruginosa. Infect. Immun. 2001, 69, 5385–5394.
- Brandel, J.; Humbert, N.; Elhabiri, M.; Schalk, I.J.; Mislin, G.L.A.; Albrecht-Gary, A.-M. Pyochelin, a siderophore of Pseudomonas aeruginosa: Physicochemical characterization of the iron(iii), copper(ii) and zinc(ii) complexes. Dalton Trans. 2012, 41, 2820–2834.
- Serino, L.; Reimmann, C.; Visca, P.; Beyeler, M.; Chiesa, V.D.; Haas, D. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 1997, 179, 248–257.
- Dumas, Z.; Ross-Gillespie, A.; Kümmerli, R. Switching between apparently redundant iron-uptake mechanisms benefits bacteria in changeable environments. Proc. R. Soc. B Biol. Sci. 2013, 280, 20131055.
- Lyczak, J.B.; Cannon, C.L.; Pier, G.B. Lung Infections Associated with Cystic Fibrosis. Clin. Microbiol. Rev. 2002, 15, 194–222.
- Lhospice, S.; Gomez, N.O.; Ouerdane, L.; Brutesco, C.; Ghssein, G.; Hajjar, C.; Liratni, A.; Wang, S.; Richaud, P.; Bleves, S.; et al. Pseudomonas aeruginosa zinc uptake in chelating environment is primarily mediated by the metallophore pseudopaline. Sci. Rep. 2017, 7, 17132.
- Ghssein, G.; Brutesco, C.; Ouerdane, L.; Fojcik, C.; Izaute, A.; Wang, S.; Hajjar, C.; Lobinski, R.; Lemaire, D.; Richaud, P.; et al. Biosynthesis of a broad-spectrum nicotianamine-like metallophore in Staphylococcus aureus. Science 2016, 352, 1105–1109.
- Skaar, E.P. The Battle for Iron between Bacterial Pathogens and Their Vertebrate Hosts. PLOS Pathog. 2010, 6, e1000949.
- Ochsner, A.U.; Vasil, I.A.; Vasil, M.L. Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression: Purification and activity on iron-regulated promoters. J. Bacteriol. 1995, 177, 7194–7201.
- Troxell, B.; Hassan, H.M. Transcriptional regulation by Ferric Uptake Regulator (Fur) in pathogenic bacteria. Front. Cell Infect. Microbiol. 2013, 3, 59.
- Totter, J.R.; Moseley, F.T. Influence of the concentration of iron on the production of fluorescin by Pseudomonas aeruginosa. J Bacteriol. 1952, 6, 45–47.
- Meyer, J.M.; Hornspreger, J. Role of pyoverdinePf the iron binding fluorescent pigment of Pseudomonas fluorescens iron transport. J. Gen. Microbiol. 1978, 107, 329–331.
- Budzikiewicz, H.; Schäfer, M.; Fernández, D.U.; Matthijs, S.; Cornelis, P. Characterization of the chromophores of pyoverdins and related siderophores by electrospray tandem mass spectrometry. BioMetals 2006, 20, 135–144.
- Boukhalfa, H.; Crumbliss, A.L. Chemical aspects of siderophore mediated iron transport. BioMetals 2002, 15, 325–339.
- Demange, P.; Wendenbaum, S.; Linget, C.; Mertz, C.; Cung, M.T.; Dell, A.; Abdallah, M.A. Bacterial siderophores: Structure and NMR assigment of pyoverdins PaA, siderophores of Pseudomonas aeruginosa ATCC 15692. Biol. Met. 1990, 3, 155–170.
- Schalk, I.J.; Guillon, L. Pyoverdine biosynthesis and secretion in Pseudomonas aeruginosa: Implications for metal homeostasis. Environ. Microbiol. 2012, 15, 1661–1673.
- Fuchs, R.; Schafer, M.; Geoffroy, V.; Meyer, J.-M. Siderotyping A Powerful Tool for the Characterization of Pyoverdines. Curr. Top. Med. Chem. 2001, 1, 31–57.
- Meyer, J.-M.; Stintzi, A.; De Vos, D.; Cornelis, P.; Tappe, R.; Taraz, K.; Budzikiewicz, H. Use of Siderophores to Type Pseudomonads: The Three Pseudomonas Aeruginosa Pyoverdine Systems. Microbiology 1997, 143, 35–43.
- Briskot, G.; Taraz, K.; Budzikiewicz, H. ChemInform Abstract: Bacterial Constituents. Part 37. Pyoverdin-Type Siderophores from Pseudomonas aeruginosa. ChemInform 1989, 1989, 375–384.
- Tappe, R.; Tara, K.; Budzikiewicz, H.; Meyer, J.M.; Lefèvre, J.F. Structure elucidation of a pyoverdine produced by Pseudomonas aeruginosa ATCC 27853. J. Prakt. Chem. 1993, 335, 83–87.
- Gipp, S.; Hahn, J.; Taraz, K.; Budzikiewicz, H. Zwei Pyoverdine aus Pseudomonas aeruginosa R./Two Pyoverdins from Pseudomonas aeruginosa R. Z. Naturforsch. C 1991, 46, 534–541.
- Ruangviriyachai, C.; Fernández, D.U.; Fuchs, R.; Meyer, J.-M.; Budzikiewicz, H. A New Pyoverdin from Pseudomonas aeruginosa R’. Z. Naturforsch. C J. Biosci. 2001, 56, 933–938.
- Takase, H.; Nitanai, H.; Hoshino, K.; Otani, T. Impact of Siderophore Production on Pseudomonas aeruginosa Infections in Immunosuppressed Mice. Infect. Immun. 2000, 68, 1834–1839.
- Potvin, E.; Sanschagrin, F.; Levesque, R.C. Sigma factors in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 2008, 32, 38–55.
- Michel, L.; Bachelard, A.; Reimmann, C. Ferripyochelin uptake genes are involved in pyochelin-mediated signalling in Pseudomonas aeruginosa. Microbiology 2007, 153, 1508–1518.
- Raymond, K.N.; Dertz, E.A.; Kim, S.S. Enterobactin: An archetype for microbial iron transport. Proc. Natl. Acad. Sci. USA 2003, 100, 3584–3588.
- Wyckoff, E.E.; Mey, A.R.; Payne, S.M. Iron acquisition in Vibrio cholerae. BioMetals 2007, 20, 405–416.
- Perraud, Q.; Cantero, P.; Roche, B.; Gasser, V.; Normant, V.P.; Kuhn, L.; Hammann, P.; Mislin, G.L.A.; Ehret-Sabatier, L.; Schalk, I.J. Phenotypic Adaption of Pseudomonas aeruginosa by Hacking Siderophores Produced by Other Microorganisms. Mol. Cell. Proteom. 2020, 19, 589–607.
- Schalk, I.J.; Cunrath, O. An overview of the biological metal uptake pathways in Pseudomonas aeruginosa. Environ. Microbiol. 2016, 18, 3227–3246.
- Gonzalez, M.R.; Ducret, V.; Leoni, S.; Perron, K. Pseudomonas aeruginosa zinc homeostasis: Key issues for an opportunistic pathogen. Biochim. Biophys. Acta 2019, 1862, 722–733.
- Fajardo, A.; Hernando-Amado, S.; Oliver, A.; Ball, G.; Filloux, A.; Martinez, J.L. Characterization of a novel Zn2+-dependent intrinsic imipenemase from Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2014, 69, 2972–2978.
- Mastropasqua, M.C.; D’Orazio, M.; Cerasi, M.; Pacello, F.; Gismondi, A.; Canini, A.; Canuti, L.; Consalvo, A.; Ciavardelli, D.; Chirullo, B.; et al. Growth of Pseudomonas aeruginosa in zinc poor environments is promoted by a nicotianamine-related metallophore. Mol. Microbiol. 2017, 106, 543–561.
- McFarlane, J.S.; Lamb, A.L. Biosynthesis of an Opine Metallophore by Pseudomonas aeruginosa. Biochemistry 2017, 56, 5967–5971.
- Laffont, C.; Brutesco, C.; Hajjar, C.; Cullia, G.; Fanelli, R.; Ouerdane, L.; Cavelier, F.; Arnoux, P. Simple rules govern the diversity of bacterial nicotianamine-like metallophores. Biochem. J. 2019, 476, 2221–2233.
- Pederick, V.G.; Eijkelkamp, B.; Begg, S.N.; Ween, M.; McAllister, L.J.; Paton, J.C.; McDevitt, C.A. ZnuA and zinc homeostasis in Pseudomonas aeruginosa. Sci. Rep. 2015, 5, 13139.
- Gi, M.; Lee, K.-M.; Kim, S.C.; Yoon, J.-H.; Yoon, S.S.; Choi, J.Y. A novel siderophore system is essential for the growth of Pseudomonas aeruginosa in airway mucus. Sci. Rep. 2015, 5, 14644.
- Zhang, Y.; Rodionov, A.D.; Gelfand, M.S.; Gladyshev, V.N. Comparative genomic analyses of nickel, cobalt and vitamin B12 utilization. BMC Genom. 2009, 10, 78.
- Crespo, A.; Pedraz, L.; Astola, J.; Torrents, E. Pseudomonas aeruginosa Exhibits Deficient Biofilm Formation in the Absence of Class II and III Ribonucleotide Reductases Due to Hindered Anaerobic Growth. Front. Microbiol. 2016, 7, 688.
- Zhang, J.; Zhao, T.; Yang, R.; Siridechakorn, I.; Wang, S.; Guo, Q.; Bai, Y.; Shen, H.C.; Lei, X. De novo synthesis, structural assignment and biological evaluation of pseudopaline, a metallophore produced by Pseudomonas aeruginosa. Chem. Sci. 2019, 10, 6635–6641.
- Gomez, N.O.; Tetard, A.; Ouerdane, L.; Laffont, C.; Brutesco, C.; Ball, G.; Lobinski, R.; Denis, Y.; Plésiat, P.; Llanes, C.; et al. Involvement of the Pseudomonas aeruginosa MexAB–OprM efflux pump in the secretion of the metallophore pseudopaline. Mol. Microbiol. 2020, 115, 84–98.
More
Information
Subjects:
Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
914
Revisions:
2 times
(View History)
Update Date:
30 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




