Subcellular mRNA localization is an evolutionarily conserved mechanism to spatially and temporally drive local translation and, in turn, protein targeting. Hence, this mechanism achieves precise control of gene expression and establishes functional and structural networks during cell growth and development as well as during stimuli response. Since its discovery in ascidian eggs, mRNA localization has been extensively studied in animal and yeast cells. mRNA localization to the endoplasmic reticulum (ER) has also been well established since its discovery in cereal endosperm cells in the early 1990s. Storage protein mRNA targeting to distinct subdomains of the ER determines efficient accumulation of the corresponding proteins in different endosomal storage sites and, in turn, underlies storage organelle biogenesis in cereal grains. The targeting process requires the presence of RNA localization elements, also called zipcodes, and specific RNA-binding proteins that recognize and bind these zipcodes and recruit other factors to mediate active transport.
1. Introduction
mRNA localization is referred to as a mechanism where mRNAs are specifically localized to discrete subcellular compartments. By creating local translation hotspots, mRNA localization provides a highly efficient process to concentrate newly synthesized proteins within a defined intracellular region. mRNA localization enables cells to fine-tune cell polarization, differentiation, and migration as well as quickly respond to intracellular and environmental stimuli.
In eukaryotic cells, localization of mRNAs involves multiple events in both the nucleus and cytoplasm. The process is initiated in the nucleus where cis-acting elements within the RNAs, also called zipcodes RNA elements, are recognized and bound by specific trans-acting factors, typically RNA-binding proteins (RBPs), to form an initial ribonucleoprotein (RNP) complex. The association of mRNAs with some of these RBPs may be maintained during RNA processing and maturation to form a primary messenger ribonucleoprotein (mRNP) complex competent for export from the nucleus. Once transported to the cytoplasm, the primary mRNP complex is remodeled by removing and recruiting one or more protein factors and linking to a cytoskeleton motor protein to initiate mRNA transport. When the mRNP complex arrives at the target location, further remodeling is required to activate local translation, turnover or storage. Thus, mRNA localization is a highly dynamic process. While zipcode elements are an essential prerequisite to determine the localization of mRNAs, multiple RBPs play an extremely important roles throughout the process.
2. mRNA Targeting to the Endoplasmic Reticulum Subdomains Is Driven by Specific RNA Zipcodes
The early view of protein translation in eukaryotes assumed that after nuclear export to the cytoplasm, mRNAs were translated at random locations within the cytosol. Proteins were localized to specific cellular compartments or organelles by peptide-based determinants. N-terminal transit peptides (TP) served as the targeting signals to the chloroplast and mitochondria, while signal peptides directed the growing nascent polypeptide chain during protein synthesis to the ER. In this latter instance, a signal recognition particle (SRP) recognizes and binds to the signal peptide of the nascent polypeptide to direct the association of a complex of mRNA, ribosome and nascent polypeptide chain to the ER membrane. Signal peptides were considered to be necessary and sufficient information to target proteins to the ER in both plant and animal cells
[1][2][3].
In rice endosperm cells, large amount of storage proteins, prolamine, glutelin and α-globulin are synthesized on the ER where these proteins are translocated to the lumen. Prolamines are retained in the lumen, where they co-assemble as intracisternal granules, which matures into an organelle labeled as ER-derived protein body-I (PB-I, prolamine)
[4]. By contrast, glutelins and α-globulins are exported from the ER lumen to the Golgi and then transported to protein storage vacuoles (PSV, containing glutelin and α-globulin) to form PB-II (
Figure 1A)
[4]. Due to the presence of signal peptide elements in these storage proteins, their synthesis on the ER and packaging of the storage proteins into PB-I and PB-II were initially thought to be dependent on their signal peptides. In 1993, using in situ hybridization at the electron microscopy level, the Okita laboratory reported that
prolamine and
glutelin mRNAs were localized on two distinct subdomains of the ER
[5].
Prolamine mRNAs were localized on the ER (PB-ER) that delimit PB-I, while
glutelin mRNAs were distributed to the adjoining cisternal ER (cis-ER) (
Figure 1A). Later, using optimized in situ RT-PCR technique, the group further discovered that when removing the translation initiation codon or signal peptide sequences, prolamine mRNAs remained targeted to the PB-ER
[6]. These studies indicate that storage protein mRNAs are not randomly localized on the ER and that the localization process is RNA-based. This hypothesis was substantiated by expressing exogenous reporter genes containing
prolamine or
glutelin RNA sequences positioned at their 3′ UTR. The
β-glucuronidase (GUS) mRNA, which by itself is normally targeted to the cis-ER, was redirected by
prolamine RNA sequences to the PB-ER
[6], while
GFP mRNAs containing
prolamine or
glutelin RNA sequences were localized to the PB-ER and cis-ER, respectively
[7].
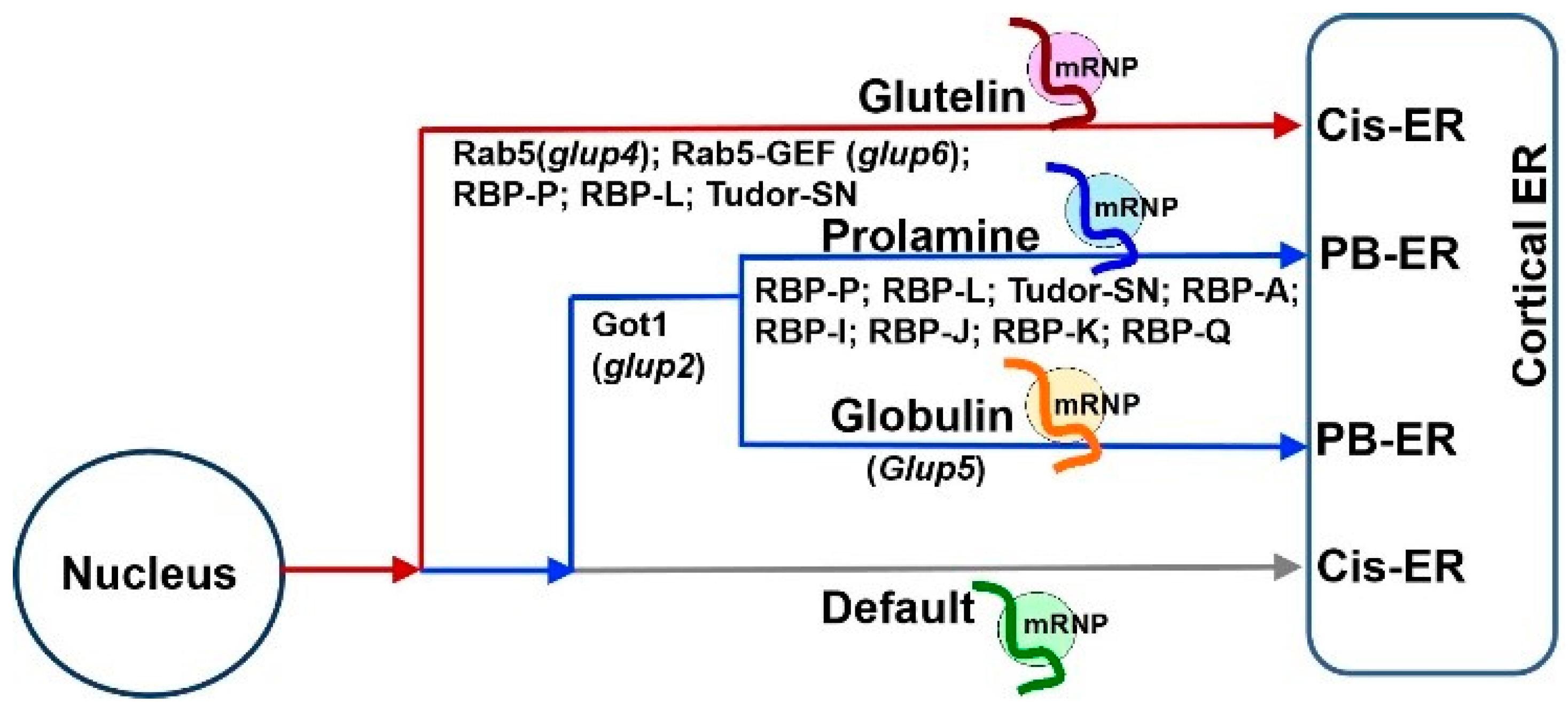
Figure 1. Schematic model of mRNA transport to the cortical ER in developing rice endosperm cells. (A) Working model of three pathways to transport mRNAs to distinct ER subdomains in wildtype. The localization of storage protein mRNAs initiates in the nucleus, where newly transcribed mRNAs are recognized and bound by various sets of specific RBPs, forming heterogenous nuclear ribonucleoprotein (hnRNP) complexes. After export from the nucleus to cytoplasm, the complexes undergo dynamic remodeling to form mRNPs that recruit molecular motor proteins for actively transporting to distinct ER subdomains likely on actin filaments (grey lines). Glutelin mRNAs (red curve line) are targeted to the cis-ER via endosomal trafficking. Following translation, proglutelins are transported via the Golgi complex to the irregularly shaped PSV, where they are proteolytically processed to acidic and basic subunits and accumulated in the crystalline regions of the PSV (shown in red). Prolamine (blue curve line) and α-globulin (orange curve line) mRNAs are transported to the PB-ER, where the mRNAs are translated and newly synthesized prolamine polypeptides are assembled as an intracisternal granule to form multi-layered PB-I. The synthesized α-globulins are rapidly exported to the Golgi for subsequent transport, via dense vesicles, to the peripheral area of the PSVs. An additional default pathway transports zipcode-less mRNAs (green curve line) to the cis-ER. (B) Mutations of key RBPs or factors induce mistargeting of storage protein mRNAs and, in turn, their proteins. Mutants carrying mutations in RBP-P, RBP-L or Tudor-SN mistarget both prolamine and glutelin mRNAs and cause changes in the shape and/or protein components of PB-I and PSVs. Got1B mutations disrupts targeting of prolamine mRNAs to the PB-ER. Due to the dysfunction of endosomal factor Rab5a and its effector Rab5a-GEF in glup6 and glup4 mutants, respectively, mRNPs (light grey dots) carrying glutelin mRNAs are partially mislocalized from the cis-ER to the PB-ER and paramural bodies (PMBs). As endosomes are also involved in glutelin and α-globulin protein trafficking from the Golgi to PSVs, both α-globulin (red dots) and proglutelin (orange dots) proteins are found to partially mislocalize in PMBs.
The direct targeting of mRNAs to the ER is best established in yeast. A set of mRNAs, including
ASH1 mRNA, are co-transported on tubular ER that move to the emerging bud or daughter cell
[8][9][10][11]. The process is found to be driven by multiple cis-acting elements within the mRNA sequences
[10]. To investigate the cis-acting elements within the storage protein mRNAs, a series of transgenic rice lines carrying a reporter gene supplemented with partial storage protein mRNA sequences was constructed
[7][12][13]. Exogenous GFP whose mRNA was localized on the cis-ER by a default pathway was used as a reporter gene to investigate
prolamine localization elements
[7]. The addition of various 5′ and 3′ deletions of the
prolamine sequences led to the identification of two apparent cis-acting elements, one located downstream of the signal peptide coding sequence (zipcode 1) and the other in the 3′ UTR (zipcode 2,
Figure 2A)
[7]. The presence of a single zipcode resulted in only partial localization of prolamine mRNA to the PB-ER. Thus, two cis-acting elements, which shared a conserved U-rich motif element (
Figure 2A), are required for restricted
prolamine mRNA localization.
Figure 2. RNA zipcodes identified in
prolamine (
A) and
glutelin (
B) mRNAs (adapted from
[14]). (
A) The proximate locations (
left) and the consensus motif sequence (
right) of the two zipcode elements in
prolamine mRNA.
Prolamine zipcode elements are located in the coding region and 3′ UTR and consists of only a single zipcode motif (*). (
B) The proximate locations (
top) and the consensus motif sequences (
bottom) of the zipcode elements in
glutelin mRNA. Glutelin mRNAs possess three zipcodes (zipcode 1, 2, 3), which consist of two motifs, zipcode motif 1 (orange triangles) and zipcode motif 2 (magenta triangles). 5′ and 3′ denote 5′ UTR and 3′ UTR, respectively.
A similar strategy was applied to identify
glutelin mRNA localization elements. When the maize
δ-zein mRNA, a member of the cereal prolamine superfamily
[15] was used as a reporter gene, two short sequences located at the 5′ and 3′ ends of the coding region as well as the 3′ UTR of
glutelin mRNA were sufficient to redirect
δ-zein mRNA from the PB-ER to the cis-ER (
Figure 2B)
[12]. These observations suggest that
glutelin mRNA contains three cis-localization elements and that the glutelin zipcodes are dominant over the
δ-zein mRNA zipcodes. Further sequence analysis suggests that these glutelin zipcode RNAs contain two conserved motifs (
Figure 2B) with the U-rich motif 2 showing some homology to the
prolamine zipcode
[12].
In addition to prolamines and glutelins, rice endosperm cells accumulate small amounts of α-globulins. The saline-soluble proteins are synthesized on the ER-membrane, processed by the Golgi and ultimately deposited together with glutelin in the PSV. Interestingly,
α-globulin mRNAs are distributed on the PB-ER (
Figure 1A), and not on the cis-ER based on the analysis of in situ RT-PCR
[13]. Its presence in the 3′ UTR of the
GFP RNA redirects the hybrid RNA from the cis-ER to the PB-ER as well
[13], suggesting that
α-globulin mRNA sequence contains cis-acting elements for localization on the PB-ER. Sequence analysis revealed that
α-globulin mRNA sequences possess three candidate zipcodes located at both the coding and non-coding regions, sharing high similarity to
prolamine zipcode RNAs
[13]. Collectively, these findings suggest that both
prolamine and
α-globulin mRNAs are directed to the cortical ER by specific zipcode RNAs.
Based on the results of these studies, three mRNA targeting pathways to the cortical ER exist in rice endosperm cells (
Figure 1A). While
prolamine and
α-globulin mRNAs are localized to the PB-ER,
glutelin mRNAs are targeted to the cis-ER. Both pathways are zipcode RNA-dependent. The third pathway is a default zipcode-independent pathway, which mediates RNAs, including
GFP and GUS mRNAs, to the cis-ER. The three pathways are not independent but instead hierarchal and inter-related (
Figure 3). The glutelin pathway is dominant, as its zipcodes can redirect the transport of
prolamine and
α-globulin mRNAs from the PB-ER to the cis-ER
[8][9] (
Figure 3). In turn,
prolamine and
α-globulin mRNAs are able to redirect
GFP RNA from the default cis-ER to the PB-ER
[7][13].
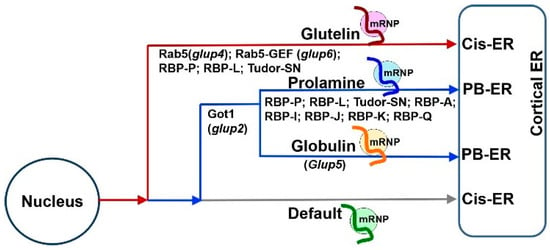
Figure 3. The hierarchal relationship among the three mRNA transport pathways in developing rice endosperm (adapted from
[16]). The
glutelin mRNA localization pathway to the cis-ER (red) is dominant over the pathway targeting
prolamine/α-globulin mRNAs to the PB-ER (blue), which, in turn, is dominant over the default pathway (gray). Studies from mutant rice lines expressing defective Got1 (
glup2), Rab5 (
glup4) and Rab5-GEF (
glup6), indicate that membrane trafficking mediates the mRNA transport to the PB-ER or cis-ER. Another rice mutant,
glup5, possessing an undefined genetic defect, misdirects α-
globulin mRNAs to the cis-ER without affecting the localization of
prolamine mRNAs to the PB-ER. The key RBPs responsible for
glutelin and
prolamine mRNA localization, are marked under each pathway. The three RNA-transport pathways may share some common RBPs or factors for mRNA targeting to the cortical ER membrane, while additional specific factors are required for selective transport during each pathway.
3. mRNA Targeting to the Endoplasmic Reticulum Subdomains Requires a Set of Trans-Acting RNA-Binding Proteins
Following the identification of cis-acting elements in RNAs, identification of transacting factors, mainly RNA-binding proteins (RBPs), was pursued. Two strategies were used to identify the RBPs required for
prolamine and
glutelin mRNA localization, respectively. Affinity chromatography using
prolamine zipcode RNA as bait to pull-down interacting RBPs was initially applied
[17]. Fifteen unique RBPs with specific binding affinity to the
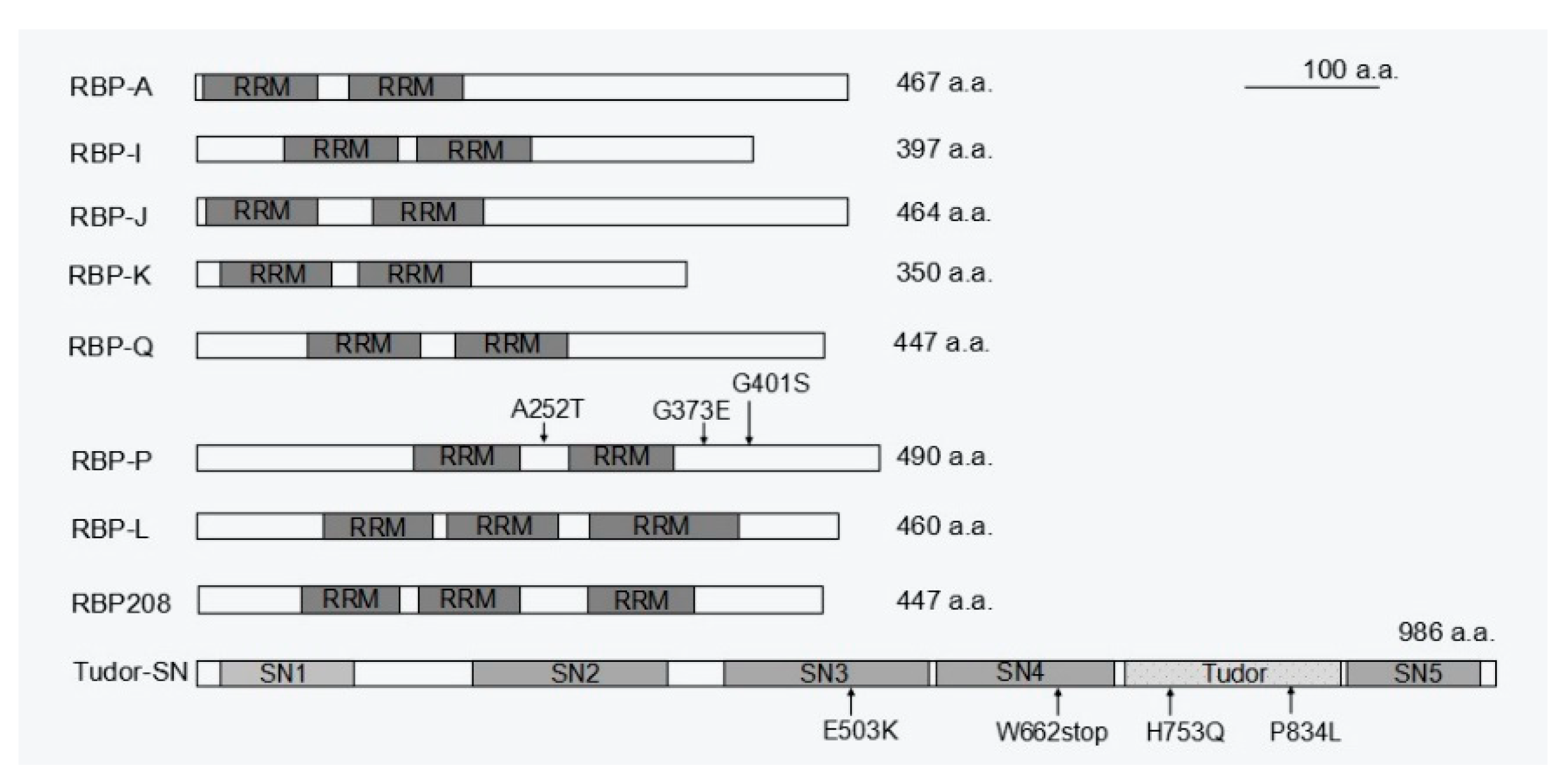
prolamine zipcode were selectively captured under highly stringent washing and elution conditions. Five of these RBPs, A, I, J, K, and Q, were heterogeneous nuclear ribonucleoproteins (hnRNPs) containing two RNA recognition motifs (RRMs) and were selected for further functional analysis
[18]. All five RBPs have binding capability to
prolamine zipcode RNAs. They form multiple complexes in the nucleus and cytoplasm, suggesting that they mediate various steps during
prolamine mRNA transport and localization (
Figure 4 and
Figure 5). RBPs A-J-K and I-J-K assemble into two complexes associated with
prolamine zipcodes in both the nucleus and cytoplasm, while RBP-Q is involved in the formation of an undefined third complex in the nucleus that is released in the cytoplasm.
Figure 4. A proposed working model of five RBPs involved in
prolamine mRNA localization (adapted from
[18]). Five RBPs, A, I, J, K, and Q assemble into at least three different RBP complexes that recognize and bind to
prolamine zipcodes. RBPs A, I, J and K form two cytoplasmic complexes, A-J-K and I-J-K. These two RBP complexes together with a third complex containing RBP-Q may also be present in the nucleus. In the nucleus, RBPs I and J may be associated with other proteins preventing their recognition by antibodies. When associated with RBP-A and RBP-K, RBP-Q is not accessible to its antibody, suggesting that it is bound by other proteins that comprise a third multiprotein family. Alternatively, the nucleus contains a simpler complex consisting of RBPs A and K as well as the RBP-Q complex.
Figure 5. Schematic structure of the available RBPs responsible for prolamine and/or glutelin mRNA localization. While RBP-A, I, J, K, Q, and P contain two RRM motifs, there are three RRMs in RBP-L and RBP208. Tudor-SN consists of four SN-like domains (SN1 to 4) followed by a Tudor domain and a fifth abbreviated SN-like domain (SN5). The mutations sites in RBP-P and Tudor-SN that cause mislocalization of prolamine and glutelin mRNAs are indicated by arrows followed by labeling amino acid substitutions. a.a., amino acids.
While affinity chromatography using prolamine zipcode RNA was successful in capturing specific
trans-RBPs, it failed to identify specific
glutelin zipcode binding proteins. An alternative North-western blot approach was later undertaken using
glutelin zipcode and non-zipcode RNA as a comparison group, which identified RBP-P as a key
glutelin zipcode-binding protein
[19]. RBP-P was also captured by
prolamine zipcode RNA chromatography
[17], suggesting a dual functional role of RBP-P in localization of both glutelin and prolamine mRNAs. Indeed, the two RRMs containing RBP-P (
Figure 5) showed highly specific binding to both
glutelin and
prolamine mRNAs, especially their zipcodes. Mutations in RBP-P led to a loss of RNA binding activity and caused partial mislocalization of both
glutelin and
prolamine mRNAs
[20]. Collectively, these results indicate RBP-P plays essential roles in mediating specific targeting of both
glutelin and
prolamine mRNAs.
Similar to the case of prolamine, multi-protein complexes are also required to determine the localization of
glutelin mRNAs. Protein–protein interaction revealed that RBP-P, RBP-L and RBP208 interact with each other, forming multiple complexes in the nucleus and/or cytoplasm
[20]. RBP-L exhibits similar features as RBP-P (
Figure 5), including binding to
glutelin and
prolamine mRNAs in vivo and in vitro
[14]. When RBP-L expression is knocked down by a DNA segmental mutation in the 3′ UTR region, both
glutelin and
prolamine mRNAs
[14] are partially mislocalized, suggesting that RBP-L is also a key RBP in regulating the localization of
glutelin and
prolamine mRNAs. Given that the complexes formed by RBP-P and RBP-L are not RNA-dependent and are located in both the nucleus and cytoplasm, the two RBPs may form a primary complex to serve as a scaffold to bind other RBPs to form a multi-protein complex that selectively targets
prolamine and
glutelin mRNAs to the cortical ER. In addition to RBP-P and RBP-L, a third candidate, RBP208, may also be involved. RBP208 interacts with RBP-P, and this interaction is weakened with several RBP-P mutant proteins. Hence, RBP208 may also be required for precise control of
glutelin and
prolamine mRNA localization. Unlike the RBP-P/RBP-L complex, RBP-P/RBP208 complexes are found in both the nucleus and cytoplasm and their interaction is RNA-dependent
[20]. Hence, RBP208 may be specially recruited by RBP-P to the complex during mRNA localization. As RBP208 only interacts with RBP-L in the cytoplasm, the RBP group P/L/208 may also form multiple complexes to co-regulate
glutelin and
prolamine mRNA localization. However, the exact detailed function of RBP208 in the process deserves further investigation.
The requirement of RBP-P, RBP-L and RBP208 for both glutelin and prolamine mRNA localization indicates that the two transport pathways are inter-related by sharing common trans-factors to assemble the required mRNP complexes. This feature further contributes to the close relationship between the glutelin and prolamine mRNA transport pathways. The current results suggest a scenario that while the primary scaffold formed by RBP-P and RBP-L, with the undefined role from RBP208, selects glutelin and prolamine mRNAs for specific targeting to the cortical ER, the complexes formed by RBPs A, I, J, K and Q are involved to target prolamine mRNAs to the PB-ER. How the RBP group A/I/J/K/Q links to RBP-P/L scaffold and what specific factors control glutelin mRNA transport pathway require further study.