Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Gul, I.; Zhai, S.; Zhong, X.; Chen, Q.; Yuan, X.; Du, Z.; Chen, Z.; Raheem, M.A.; Deng, L.; Leeansyah, E.; et al. Angiotensin-Converting Enzyme 2-Based Biosensing Modalities. Encyclopedia. Available online: https://encyclopedia.pub/entry/36121 (accessed on 07 February 2026).
Gul I, Zhai S, Zhong X, Chen Q, Yuan X, Du Z, et al. Angiotensin-Converting Enzyme 2-Based Biosensing Modalities. Encyclopedia. Available at: https://encyclopedia.pub/entry/36121. Accessed February 07, 2026.
Gul, Ijaz, Shiyao Zhai, Xiaoyun Zhong, Qun Chen, Xi Yuan, Zhicheng Du, Zhenglin Chen, Muhammad Akmal Raheem, Lin Deng, Edwin Leeansyah, et al. "Angiotensin-Converting Enzyme 2-Based Biosensing Modalities" Encyclopedia, https://encyclopedia.pub/entry/36121 (accessed February 07, 2026).
Gul, I., Zhai, S., Zhong, X., Chen, Q., Yuan, X., Du, Z., Chen, Z., Raheem, M.A., Deng, L., Leeansyah, E., Zhang, C., Yu, D., & Qin, P. (2022, November 23). Angiotensin-Converting Enzyme 2-Based Biosensing Modalities. In Encyclopedia. https://encyclopedia.pub/entry/36121
Gul, Ijaz, et al. "Angiotensin-Converting Enzyme 2-Based Biosensing Modalities." Encyclopedia. Web. 23 November, 2022.
Copy Citation
Rapid and cost-effective diagnostic tests for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are a critical and valuable weapon for the coronavirus disease 2019 (COVID-19) pandemic response. SARS-CoV-2 invasion is primarily mediated by human angiotensin-converting enzyme 2 (hACE2). Recent developments in ACE2-based SARS-CoV-2 detection modalities accentuate the potential of this natural host-virus interaction for developing point-of-care (POC) COVID-19 diagnostic systems.
COVID-19
ACE2 biosensors
SARS-CoV-2
electrochemical detection
1. Introduction
Coronavirus disease 2019 (COVID-19) pandemic is still ongoing, posing a severe threat to human health and global healthcare system. The development of on-demand, rapid and cost-efficient severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) detection systems is critical to control the unabated pandemic. Although many COVID-19 sensing systems have been developed so far [1], each approach has its pros and cons. Point-of-care testing (POCT) devices are of paramount importance for both resource rich and constrained settings [2]. Introducing a biosensing approach that meets the “ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free and deliverable to end users)” criteria defined by the world health organization (WHO) [3] always remains a prime focus of a biosensing study. Rapid and low-cost diagnostic tests could revolutionize the infectious disease diagnostics by enabling the real-time pathogen detection at population level and assist the government in making timely decisions to curb disease propagation [4]. Although many COVID-9 sensing tools are commercially available, the commercialized methods have various limitations, such as low sensitivity (antigen- and antibody-based testing) and prolonged assay time (nucleic acid testing) [5]. Therefore, novel detection approaches intended to meet ASSURED criteria for SARS-CoV-2 are need of the time.
Based on bioanalyte type, SARS-CoV-2 diagnostics can be categorized into three modalities: (i) nucleic acid detection, (ii) serology testing, and (iii) imaging-based detection (computed tomography) [1]. Polymerase chain reaction (PCR) is a gold standard for COVID-19 diagnosis [6]. Several PCR-based methods have been reported for COVID-19 diagnosis [6]. The limit of detection (LOD) of most of the PCR-based assays is about ≤ 100 copies per milliliter [7], that corresponds to a viral load equal to 2–3 days before the onset of disease symptoms [8]. Among PCR variants, the RT-PCR is a widely used platform for SARS-CoV-2 detection. More than 350 SARS-CoV-2 RT-PCR kits are commercially available [9]. Garg et al. [9] compared the performance of seven commercial RT-PCR tests, including Black Bio’s TRUPCR SARS-CoV-2, Seegene’s Allplex 2019-nCOV test, Lab Gun COVID-19 RT-PCR kit from Lab Genomics, Korea, My Lab’s Patho detect COVID-19 kit, TaqPath RT-PCR COVID-19 kit from Thermo Fisher Scientific, Fosun RT-PCR kit (Fosun Ltd.), and BGI’s real-time RT-PCR kit. The analysis showed that the performance (sensitivity, specificity, and positive and negative predictive values) of BGI, Black Bio, Thermo Fisher Scientific, and Seegene are 100.0%. For weakly positive samples, the diagnostic efficacy of Lab Gun, Fosun, and My Lab is relatively lower. For a detailed account of the performance of SARS-CoV-2 sensing systems, readers are referred to a recent review dedicated to comparative analysis of SARS-CoV-2 diagnostics [7]. However, PCR-based sensing is costly, labor-intensive, and demands experienced users. To overcome these limitations, efforts have been made to develop amplification-free on-demand detection systems [10][11][12]. Unfortunately, the majority of the amplification-free SARS-CoV-2 detection approaches are unable to achieve clinically useful sensitivity for real-sample analysis and continue to recommend nucleic acid pre-amplification prior to testing [13][14], resulting in an increased assay time and cost.
In addition to PCR-based COVID-19 diagnostics, lateral flow assays (LFAs) have shown increased research trend due to certain features, such as rapid response, low-cost, and suitability for both resource-rich and resource-constrained settings. Based on target detection, LFAs are categorized into three types: nucleic acid-, antibodies-, and antigen-based LFAs. According to Global Market Insights (GMI) [15], the LFA market size exceeded USD 7.9 billion in 2021, and a compound annual growth rate (CGAR) of over 5.7% between 2022 and 2028 (>$12.1 billion in 2028) is projected. Commercial LFA products have variable efficiencies depending on the assay conditions. The sensitivities and specificities of the LFAs range from 57.6–100% and 95.87–100%, respectively [16]. A recent study on evaluation of the diagnostic performance of two commercial unsupervised rapid antigen tests reports the sensitivities of 65.7% and 62.1%, while the specificities are 100% [17]. In addition to the signal transduction system used in a detection method, the biocomponent plays a pivotal role in accuracy of the proposed or developed approach. In this regard, SARS-CoV-2 entry receptor Human angiotensin-converting enzyme 2 (hACE2) has enticed research attention as a potential biocomponent.
hACE2 is a well-studied drug target for SARS-CoV-2 treatment [18][19][20]. In early COVID-19 controlling strategies, many studies suggested the possibility of repurposing the drugs effective for other SARS coronaviruses to treat SARS-CoV-2, which could decrease the cost and development time of SARS-CoV-2 therapeutics. The spike proteins of SARS-CoV-1 and SARS-CoV-2 have a sequence similarity of 76% [21], and receptor-binding domains of SARS-CoV-1 and SARS-CoV-2 have an overall sequence similarity of 73–76% [22]. Despite this similarity, a little efficacy against SARS-CoV-2 has been demonstrated in hospitalized COVID-19 cases for SARS-CoV-1 drugs [23]. Several therapeutic approaches targeting ACE2 against COVID-19 have been reported [24], and the virus’s entry into host cells is studied using atomic force microscopy (AFM) [25].
Commonly used SARS-CoV-2 detection methods are summarized in Figure 1.
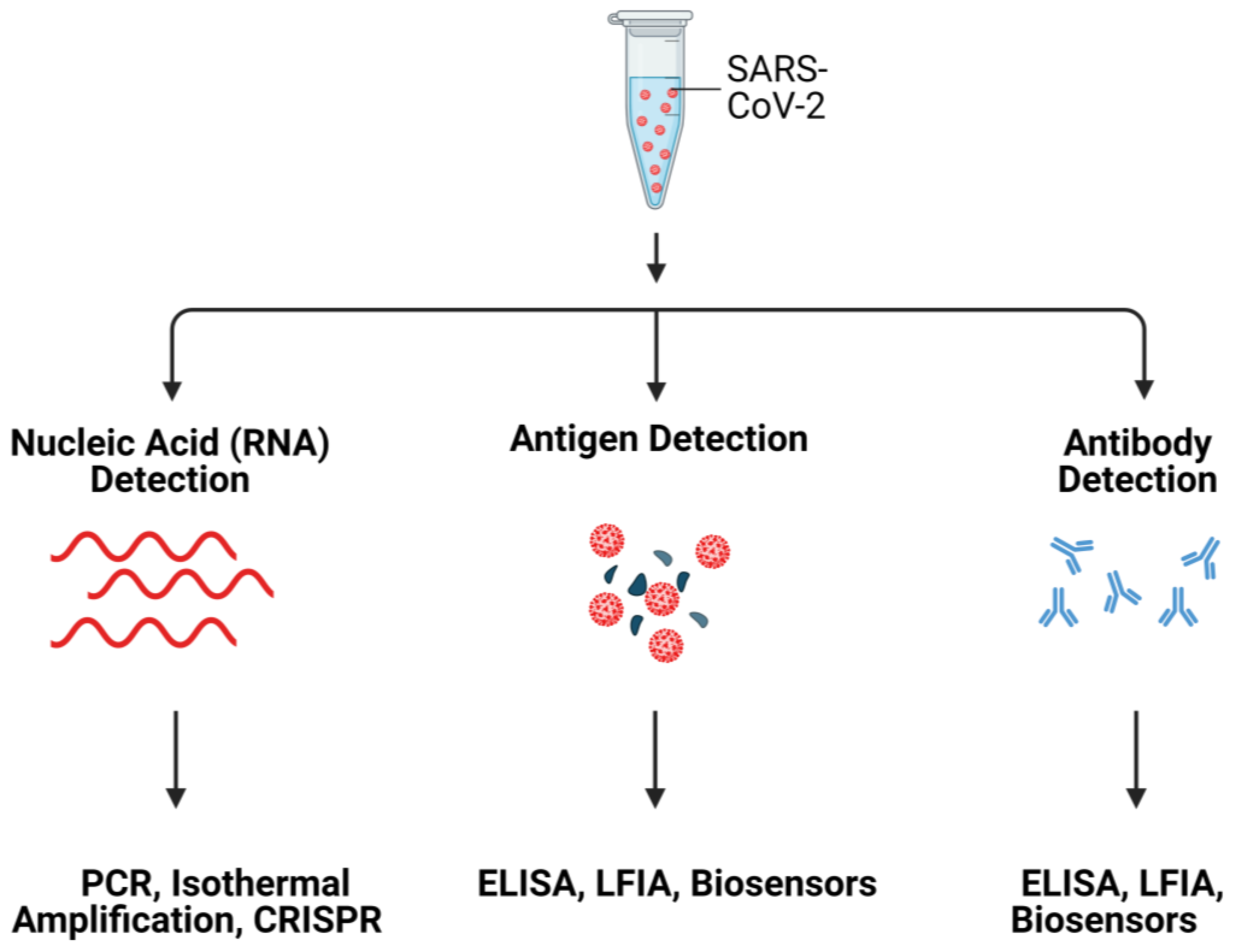
Figure 1. Schematic illustration of SARS-CoV-2 detection systems. Created with Biorender.
2. ACE2: A Mediator for SARS-CoV-2 Invasion into Host Cells
ACE2 is a carboxypeptidase that hydrolyses a single amino acid from the C-terminus of the substrate. ACE2 is the homolog of ACE with 40% amino acid sequence identity. However, the substrate specificity and cleavage pattern of ACE2 are different from those of ACE, a dipeptidase [26]. ACE2 has multiple physiological functions including, catalytic activities with peptide substrates, receptor for SARS-CoVs [27], and amino acid transporter [28]. ACE2 is involved in peptidergic renin-angiotensin system (RAS) that functions in the homeostatic control of cardiovascular and renal systems as well as regulation of extracellular fluid volume [29]. In RAS, the ACE2 converts angiotensin I (ang I) into angiotensin-(1–9) and angiotensin II (ang II) into angiotensin-(1–7) [28][30], where ACE2 plays important roles in the onset and development of myocardial infarction(MI) [29], diabetic cardiovascular complications, systemic and pulmonary hypertension, and COVID-19 pathogenesis [30].
The open reading frame of human ACE2 encodes an 805-residue polypeptide. The full-length ACE2 has two domains: a protease catalytic domain (PD) comprising of 598 residues and a collectrin-like domain (CLD) with 190 residues [31]. The PD is present at the N-terminus where a 17-residue signal peptide (SP) is also located and is removed during post-translational modifications. The key sites present in PD include an active site (for peptidase activity), a spike-RBD binding site, a hinge region back of the active site, and a PD dimerization interface (Figure 2) [31]. The hinge region is involved in the movement of the active site, leading to an “open,” “close,” or “intermediate” conformations of the PD. This indicates that the active site and hinge pocket are allosterically linked. Further, spike protein binding to ACE2 increases catalytic activity of the enzyme, which supports the notion that the RBD binding site of ACE2 might be regulated allosterically [32][33]. The CLD is present at the C-terminus and is mainly involved in ACE2 membrane anchorage [31]. The CLD is comprised of a neck domain that is involved in ACE2 dimerization, a cleavage region, a transmembrane helix, and a cytosolic tail.
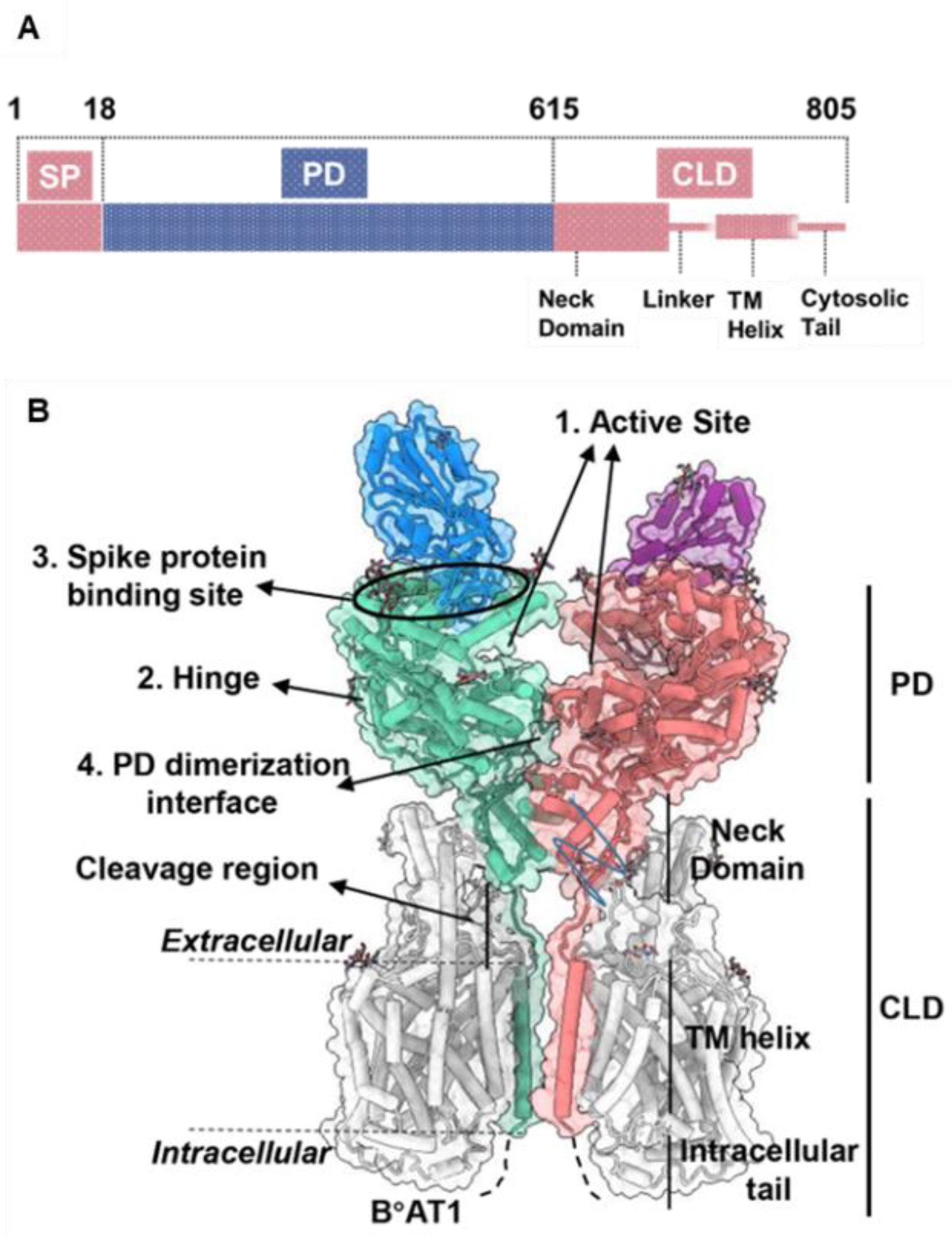
Figure 2. ACE2 gene domain organization and protein structure. (A), Full-length ACE2 containing a signal peptide (SP), an N-terminal peptidase domain (PD), and a C-terminal collectrin-like domain (CLD). (B), Protein structure of full-length ACE2 dimer in complex with B°AT1 (PDB ID: PDB 6M17). B°AT1 is presented in grey and four key sites (1–4) are highlighted. The numbers (1–805) indicate the amino acid residues.
The full-length ACE2 is a dimer [25]. The dimeric soluble ACE2 (PD domain of the full-length ACE2) shows higher affinity for the S trimer compared to the monomeric soluble ACE2 [34]. In an effort to enhance the S-binding affinity of the soluble ACE2 to use it as a decoy receptor, an ACE2 trimer is created using the foldon tag of bacteriophage T4 fibritin [34]. Due to the fact that all of the ACE2 monomers in the trimeric ACE2 bind to S trimer, the newly engineered ACE2 trimer has a stronger affinity for S trimer. The newly engineered ACE2 trimer showed an enhanced affinity for S trimer due to the involvement of all ACE2 monomers (in the trimeric ACE2) in binding to S trimer.
The identification of the disease-specific biomarkers is the first step towards development of a biosensing system. SARS-CoV-2 is an enveloped, positive-sense single-stranded RNA virus, belonging to the family Coronaviridae [35][36]. SARS-CoV-2 has four structural proteins; spike protein (S), envelope protein (E), membrane protein (M), and the nucleocapsid protein (N)] [36][37], approximately sixteen non-structural proteins and 5–8 accessory proteins [35]. The spike, membrane and envelope proteins (glycoproteins) are present on the outer part of the virus, whereas nucleocapsid protein encapsulates viral genome. The M protein is responsible for virus shape, the E protein is involved in virion assemble and release while the N protein packages the viral genome [38]. The S protein of coronavirus has two functional subunits: S1 and S2. The re ceptor-binding domain (RBD) is present in S1 subunit while S2 subunit mediates the fusion of the host-virus membranes [39]. Schematics of the AFM studies elucidating the molecular mechanism of SARS-CoV-2 binding to the host cell receptor (Figure 3) [39]. The S protein interacts with the ACE2 leading to the virus tight binding with the host cell surface, followed by virus internalization into the host cell. SARS-CoV-2 enters host cells via two distinct pathways, endosomal entry and cell surface entry [40]. In both pathways, the S protein cleavage is a prerequisite for viral entry, and involves two S protein cleavage events. One cleavage occurs at the S1-S2 subunit junction of S protein, while the other occurs at the S2′ site, which lies internal to the S2 subunit. The cleavage of the S1-S2 junction occurs during virus maturation in the infected cells. The cleavage of S2′ leads to the fusion of viral and cellular membranes, resulting in the release of viral RNA into the cytoplasm. Upon binding to ACE2, if the virus-ACE2 complex does not encounter TMPRSS2 or if there is a low expression of TMPRSS2 on the host cell surface, the SARS-CoV-2 is internalized by endocytosis. After endocytosis, the S2′ is cleaved by cathepsins in an acidic environment, which exposes the fusion peptide (FP). In both entry pathways, the S2′ exposes the FP. The conformational changes in S2 resulting from the dissociation of S1 from S2 result in orientational changes in FP, bringing it closer to the target membrane and leading to the release of viral RNA into the host cytoplasm for replication [40].
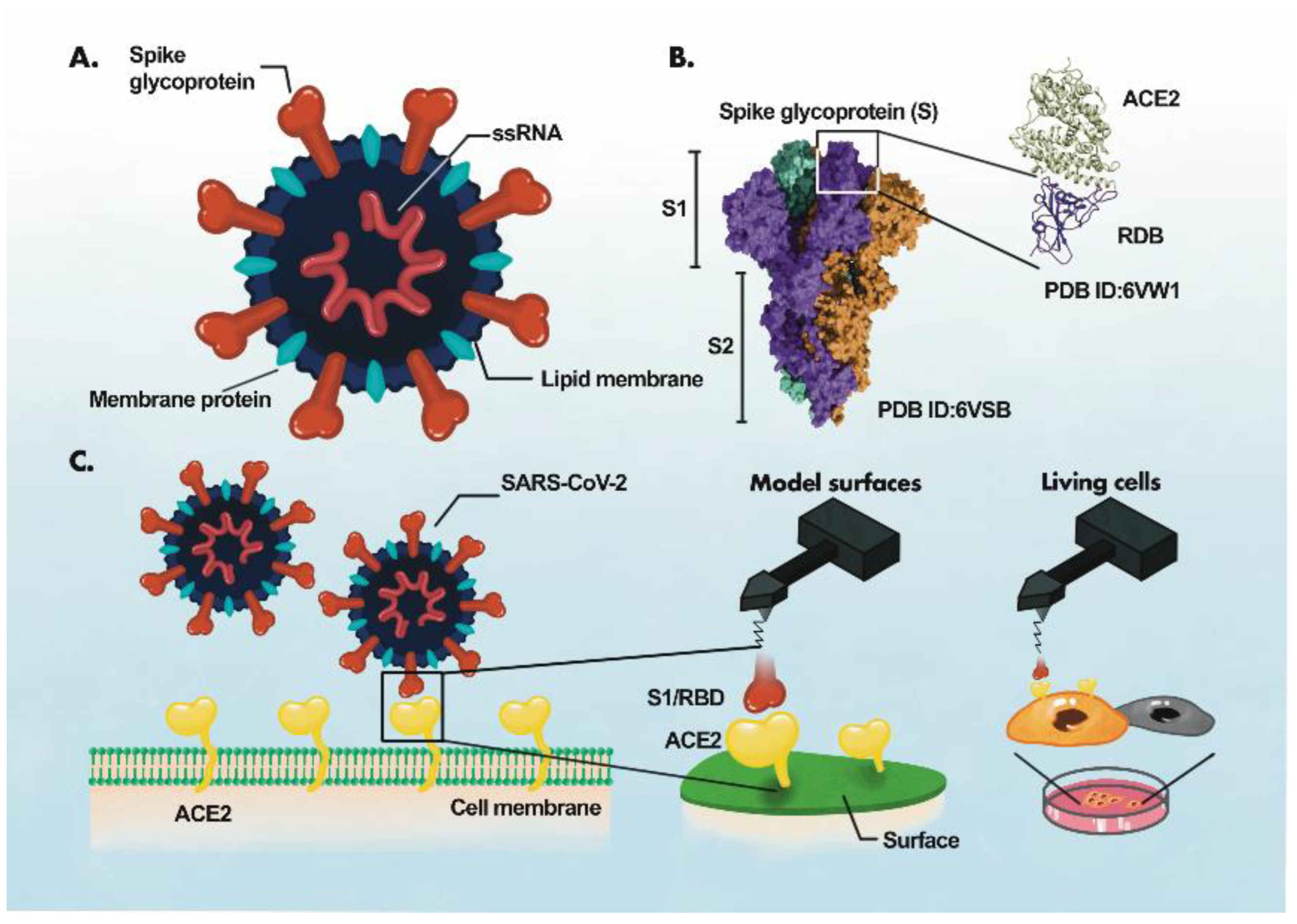
Figure 3. AFM study of ACE2-S- protein binding mechanism. (A), Components of the virus particle. (B), Complex between receptor-binding domain (RBD) of SARS-CoV-2 and ACE2. (C), Atomic force microscopy (AFM) study of the ACE2 binding with RBD. The ACE2 receptor is immobilized on a surface and S1 subunit or RBD is attached to the surface of the AFM tip.
3. Harnessing ACE2 as a Bioreceptor
COVID-19 pandemic has attracted researchers from different fields to develop rapid and accurate virus detection systems for mitigation of disease progression in a timely fashion. Despite this surge in COVID-19 biosensing, the developed methods still lack the complete characteristics of an ideal detection system. An ideal biosensing approach should meet the following prerequisites: multiplex detection, high sensitivity for real-sample analysis, high selectivity, rapid response time, portability, disposability, prolonged shelf life, cost-effectiveness, mass manufacturability, and ease of use [38][42][43][44]. Incorporating these features into a new biosensing method or technology always remains the ultimate goal of future detection platforms. In this regard, SARS-CoV-2 biosensors have shown increased research trend since the onset of COVID-19. A biosensor is an integrated device comprising of three components: (i) biorecognition element (bioreceptor); (ii) signal transducer; and (iii) signal detector [45][46]. The interaction between biocomponent and bioanalyte is recognized by the transducer and is translated into a readable signal using digital devices such as embedded devices and smartphones. Biosensors are classified mainly based on the bioreceptor and signal transduction mechanism [46][47]. The use of ACE2 as a bioreceptor for COVID-19 biosensors seems appealing due to certain advantages of ACE2 over other bioreceptors. For instance, a computational analysis shows that the mutations in S-RBD improve the binding of S protein with ACE2, leading to enhanced infectivity [48]. These mutations can lead to false negative results in the case of nucleic acid testing. Since ACE2 is a natural receptor, in contrast to other biorecognition elements, no modifications to ACE2 are needed for variant detection. This ACE2-S protein natural interaction led to the development of ACE2 biosensors for SARS-CoV-2 variants [49][50]. Further, an ACE2 biosensor also detects neutralizing antibodies [50], showing its broad application potential. The amino acid sequence diversity of spike protein among coronaviruses allows for ACE2-S-based SARS-CoV-2 biosensing. The ACE2 can be used as a substitute for the capture antibody to selectively bind the virus, resulting in an ACE2-based immunodetection system where a widely used enzyme linked immunosorbent assay (ELISA) can be easily repurposed into a novel virus assay. Colorimetric detection is a user-friendly approach amenable to automation and realization in POC devices. Similarly, electrochemical detection has garnered significant research attention for the development of POCTs. The binding of ACE2 with the spike protein of the virus may result in an electrochemical event that can sensitively sense the presence of virus [51].
4. ACE2-Based State-of-the-Art SARS-CoV-2 Detection
The ACE2-virus binding event can be translated into different signal outputs depending on the type of signal transduction system used in the experiment (Figure 4).
4.1. ACE2-Based Electrochemical Detection of SARS-CoV-2
The electrochemical behavior of the bioreceptor-bioanalyte interaction can be translated into a sensitive electrochemical biosensor. The electrical methods explored for SARS-CoV-2 detection include potentiometry [55], amperometry [56], electrochemical impedance spectroscopy (EIS) [57], differential pulse voltammetry [58][59], and square wave voltammetry [60]. In the amperometric technique, the current generated by the electroactive species is measured and correlated to the analyte concentration [61]. In voltammetry, the potential difference is measured between the working electrode and the reference electrode. The accumulation of a charge potential at the working electrode is measured compared to the reference electrode in an electrochemical cell when zero or no significant current flows between them [58][61]. The bioreceptor-bioanalyte interaction may lead to the change in conductance of the electrolyte solution and can be leveraged as a biosensing signal for developing an impedimetric or conductometric biosensor [62]. In electrochemical biosensors, the electrode is one of the important components. Since the bioelectrochemical event is generally monitored in the close proximity of the working electrode surface, the electrode functionalization with conductive materials and efficient immobilization of the bioreceptor are immensely important to achieve the desirable performance of the biosensor.
An electrochemical SARS-CoV-2 biosensor is developed by Torres et al. [51]. The working electrode is functionalized by the drop-casting method. The ACE2 immobilization on the working electrode is achieved by glutaraldehyde cross-linking. Bovine serum albumin (BSA) is used to avoid non-specific adsorption on the electrode surface. Electrode functionalization is systematically optimized and each fictionalization step is evaluated by cyclic voltammetry and EIS. The CV and Nyquist plots showed a successful functionalization of the working electrode. A miniaturized handheld device called RAPID (Real-time Accurate Portable Impedimetric Detection) is developed. The binding of the target analyte with ACE2 causes a change in interfacial electron transfer kinetics between the redox probe in solution and conducting electrode sites. Electrochemical impedance spectroscopy (EIS) is employed to detect the sensing signal. The calibration curves obtained with two different samples (spike protein and inactivated virus) show a satisfactory sensitivity and robustness of the method. The use of Nafion as a protective layer (and high concentration of ACE2) contribute to the sensitivity and robustness of the biosensor. The sensitivity and specificity of the RAPID for nasopharyngeal/oropharyngeal swabs and saliva samples are 85.3% and 100%, and 100% and 86.5% respectively, with an assay volume of 10 µL and a response time of 4 min. The detection limit of the biosensor is 2.18 fg mL−1 (in buffer) and 1.39 pg mL−1 for saliva samples.
In another development [24], the palladium nano thin film electrode (Pd-NTF) is fabricated and functionalized by exploiting Pd-S bond formation between the active Pd surface and ACE2. 1-octadecanethiol is used as blocking agent. The functionalized electrode is subsequently coupled to the EIS to realize a complete biosensor. The practical utility of the platform is demonstrated by screening modulators of SARS-CoV-2 S-protein-ACE2 interaction using a very small assay volume. By using the developed platform, potential pharmacological therapy that leads to suppressed virus binding with ACE2 is also identified.
ACE2 has two hydrophobic regions, the 18-amino-acid signal peptide on the N-terminus and a 22-amino-acid region on the C-terminus of the enzyme. The hydrophobic region facilitates its anchoring to the cell membrane [63] and allows ACE2 immobilization on amphiphobic structures resembling to cell membrane. Fluorous self-assembled monolayers (SAM) have been used in thin-film transistors (TFTs) to reduce biofouling of material surface [64]. Based on antibiofouling behavior and possibility of effective enzyme immobilization on electrode surface, Vezza et al., [65] deploy a perfluorocarbon self-assembled monolayer of 1H,1H,2H,2H-Perfluorodecanethiol (PFDT) on gold sensor surface to form an amphiphobic surface where ACE2 is immobilized by physisorption on PFDT. The sensor is challenged to the streptavidin, interleukin 6 (IL6), viral transport medium, and saliva samples to test its specificity. The biosensor shows clinically relevant analytical properties.
A low-cost biosensor, LEAD (Low-cost Electrochemical Advanced Diagnostic) composed of pencil graphite electrodes, is developed by de Lima et al. [52]. The transducer is modified with gold nanoparticles (AuNPs), cysteamine (cys), and bovine serum albumin (BSA) by using different cross-linkers. The ACE2 is anchored on the modified electrode, resulting in the ACE2-AuNPs-cys. The use of gold nanoparticles increases the electrode’s sensitivity due to efficient electron transfer between the electrode and the redox probe ([Fe(CN)6]−3/−4). The biosensor could detect 229 fg mL−1 spike protein. Further, the practical utility of the system is demonstrated by detecting virus in clinical samples. Moreover, the biosensor is insensitive to other potential interfering viruses with shelf-life of 5 days. Rapid response time and the use of easily accessible and commercially available materials are the key attributes of the developed biosensor.
4.2. ACE2-Based Optodiagnostics for COVID-19
Immunodetection systems have garnered substantial research attention due to their certain features, such as rapid response, ease of fabrication, and low cost. The use of an appropriate antibody pair for SARS-CoV-2 detection may improve the sensitivity and reliability of the system. In this regard, a rapid SARS-CoV-2 detection method based on ACE2 is developed [53], where ACE2 could form a matched pair with commercial antibodies. The ACE2 is used as a capture probe to selectively bind the SARS-CoV-2 S1 subunit of the S protein. Three different commercially available antibodies (CR3022, F26G19, and S1-mAb) are systematically evaluated to find the best pair with ACE2. Interestingly, the developed assay is more sensitive to the RBD than the S1, indicating the enhanced affinity of ACE2 for RBD. Spike proteins from other related viruses, such as SARS-CoV-1 and Middle East respiratory syndrome coronavirus (MERS-CoV) show no cross-reactivity. The LOD of the developed system is 1.86 × 105 copies per milliliter, and this is the first study to detect COVID-19 antigen in a lateral flow format using ACE2 as a capture probe. The study opens avenues to develop mutation-free virus detection assays where bioreceptor-virus interaction can be realized as a detection approach. A förster resonance energy transfer (FRET) phenomenon is exploited to develop a SARS-CoV-2-derived RBD ACE2 biosensor [66]. The FRET probe is developed by labelling RBD with YPet and Turquoise2-GL as acceptor and donor, respectively. The biosensor could sensitively detect the interaction between hACE2 and the RBD fragment derived from the SARS-CoV-2 spike protein, demonstrating its potential application in drug design.
In another development [67], a smartphone-assisted ACE2-based optodiagnostic system is designed and developed. Gold nanoparticles and cysteamine are used [52] and fabricated into a user-friendly sensing system. The cotton swab is functionalized with ACE2, and ACE2 is immobilized on the AuNP-cys. The ACE2-functionalized cotton swab is dipped into the sample where the analyte (spike protein or virus particle) is captured by the ACE2. After that, AuNP-cys-ACE2 is added to the reaction system and a color change is observed. The color change is attributed to the binding of virus/spike protein with ACE2, resulting in a sandwich formation that leads to plasmonic change in the AuNPs. The analytical features of the assay are comparable to the other related reports [68][69]. Prolonged incubation (>5 min) of the assay enabled adsorption of the functionalized nanoparticles onto cotton swabs, resulting in a false positive result. The use of blocking agents or a relatively inert support material may decrease the unwanted interactions of the functionalized AuNPs.
4.3. SERS-Based Detection
The development timeline of COVID-19 detection technologies indicates that most of the reported methods are particularly focused on SARS-CoV-2 detection in biological fluids. Waste water-based epidemiology (WWE) has been employed to detect different biomarkers, such as pharmaceuticals, drugs, industrial chemicals, and biologicals in wastewater [44]. Human viruses have been detected in wastewater [70][71]. The proper monitoring of viruses in environmental water can help in pandemic preparedness [72]. For example, SARS-CoV-2 RNA was detected in sewage waters in the Netherlands [73] and Australia [70]. The copy number per mL significantly increased with the increasing prevalence of the virus in the population, indicating the potential of sewage water surveillance as a promising tool to monitor the virus load in a population. The discovery of the enhancement of Raman scattering induced by molecules adsorbed on the surface of nanostructured metals has revolutionized the analytical techniques to achieve desired sensitivities. In the surface-enhanced Raman scattering (SERS) phenomenon, the light scattering is greatly enhanced by molecules adsorbed onto the corrugated metallic surfaces, leading to single-molecule SERS sensing [74]. The approach has demonstrated the single-molecule sensitivity [75][76][77].
In an effort to detect SARS-CoV-2 in environmental samples and harness the latent potential of SERS technology to combat the COVID-19 pandemic, a SERS sensor is designed and developed by Yang et al. [54]. The sensor, hACE2 immobilized gold-nanoneedle arrays (GNAs), shows a 106-fold virus enrichment attributed to the strong ACE2 binding affinity for SARS-CoV-2 and the unique geometry of the GNAs obstructing virus escape. The strong SARS-CoV-2 binding affinity of the ACE2 and the oblique hierarchical structure of the GNAs, and electromagnetic field effect coupling, synergistically result in a 109-fold enhancement of the SERS signal and a detection limit of 80 copies mL−1. The biosensor integration with machine-learning methods (principal component analysis and discrimination analysis) reliably differentiates the SARS-CoV-2 SERS signal from that of SARS-CoV-1. The practical application of the system is demonstrated by using urine samples spiked with virus, indicating the potential of SERS sensor for virus detection in wastewater.
References
- Han, T.; Cong, H.; Shen, Y.; Yu, B. Recent Advances in Detection Technologies for COVID-19. Talanta 2021, 233, 122609.
- Manmana, Y.; Kubo, T.; Otsuka, K. Recent Developments of Point-of-Care (POC) Testing Platform for Biomolecules. TrAC Trends Anal. Chem. 2021, 135, 116160.
- Deroco, P.B.; Wachholz Junior, D.; Kubota, L.T. Recent Advances in Point-of-Care Biosensors for the Diagnosis of Neglected Tropical Diseases. Sens. Actuators B Chem. 2021, 349, 130821.
- Valera, E.; Jankelow, A.; Lim, J.; Kindratenko, V.; Ganguli, A.; White, K.; Kumar, J.; Bashir, R. COVID-19 Point-of-Care Diagnostics: Present and Future. ACS Nano 2021, 15, 7899–7906.
- Rezvani Ghomi, E.; Khosravi, F.; Mohseni-M, A.; Nourbakhsh, N.; Haji Mohammad Hoseini, M.; Singh, S.; Hedenqvist, M.S.; Ramakrishna, S. A Collection of the Novel Coronavirus (COVID-19) Detection Assays, Issues, and Challenges. Heliyon 2021, 7, e07247.
- Ilkhani, H.; Hedayat, N.; Farhad, S. Novel Approaches for Rapid Detection of COVID-19 during the Pandemic: A Review. Anal. Biochem. 2021, 634, 114362.
- Filchakova, O.; Dossym, D.; Ilyas, A.; Kuanysheva, T.; Abdizhamil, A.; Bukasov, R. Review of COVID-19 Testing and Diagnostic Methods. Talanta 2022, 244, 123409.
- Ejima, K.; Kim, K.S.; Ludema, C.; Bento, A.I.; Iwanami, S.; Fujita, Y.; Ohashi, H.; Koizumi, Y.; Watashi, K.; Aihara, K.; et al. Estimation of the Incubation Period of COVID-19 Using Viral Load Data. Epidemics 2021, 35, 100454.
- Garg, A.; Ghoshal, U.; Patel, S.S.; Singh, D.V.; Arya, A.K.; Vasanth, S.; Pandey, A.; Srivastava, N. Evaluation of Seven Commercial RT-PCR Kits for COVID-19 Testing in Pooled Clinical Specimens. J. Med. Virol. 2021, 93, 2281–2286.
- Kashefi-Kheyrabadi, L.; Nguyen, H.V.; Go, A.; Baek, C.; Jang, N.; Lee, J.M.; Cho, N.-H.; Min, J.; Lee, M.-H. Rapid, Multiplexed, and Nucleic Acid Amplification-Free Detection of SARS-CoV-2 RNA Using an Electrochemical Biosensor. Biosens. Bioelectron. 2022, 195, 113649.
- Sampad, M.J.N.; Zhang, H.; Yuzvinsky, T.D.; Stott, M.A.; Hawkins, A.R.; Schmidt, H. Optical Trapping Assisted Label-Free and Amplification-Free Detection of SARS-CoV-2 RNAs with an Optofluidic Nanopore Sensor. Biosens. Bioelectron. 2021, 194, 113588.
- He, Q.; Chen, Q.; Li, F.; Yuan, X.; Wang, C.; Liu, C.; Xu, L.; Zhong, X.; Wei, J.; Pandey, V.; et al. Amplification-Free Detection of Viral RNA by Super Resolution Imaging-Based CRISPR/Cas13a System. bioRxiv 2021.
- Ma, L.; Yin, L.; Li, X.; Chen, S.; Peng, L.; Liu, G.; Ye, S.; Zhang, W.; Man, S. A Smartphone-Based Visual Biosensor for CRISPR-Cas Powered SARS-CoV-2 Diagnostics. Biosens. Bioelectron. 2022, 195, 113646.
- Sridhara, S.; Goswami, H.N.; Whyms, C.; Dennis, J.H.; Li, H. Virus Detection via Programmable Type III-A CRISPR-Cas Systems. Nat. Commun. 2021, 12, 5653.
- Global Market Insights Lateral Flow Assay Market Size by Products. Available online: https://www.gminsights.com/industry-analysis/lateral-flow-assays-market (accessed on 15 July 2022).
- Zhou, Y.; Wu, Y.; Ding, L.; Huang, X.; Xiong, Y. Point-of-Care COVID-19 Diagnostics Powered by Lateral Flow Assay. TrAC Trends Anal. Chem. 2021, 145, 116452.
- Møller, I.J.B.; Utke, A.R.; Rysgaard, U.K.; Østergaard, L.J.; Jespersen, S. Diagnostic Performance, User Acceptability, and Safety of Unsupervised SARS-CoV-2 Rapid Antigen-Detecting Tests Performed at Home. Int. J. Infect. Dis. 2022, 116, 358–364.
- Mahdian, S.; Zarrabi, M.; Panahi, Y.; Dabbagh, S. Repurposing FDA-Approved Drugs to Fight COVID-19 Using in Silico Methods: Targeting SARS-CoV-2 RdRp Enzyme and Host Cell Receptors (ACE2, CD147) through Virtual Screening and Molecular Dynamic Simulations. Inform. Med. Unlocked 2021, 23, 100541.
- Pokhrel, S.; Bouback, T.A.; Samad, A.; Nur, S.M.; Alam, R.; Abdullah-Al-Mamun, M.; Nain, Z.; Imon, R.R.; Talukder, M.E.K.; Tareq, M.M.I.; et al. Spike Protein Recognizer Receptor ACE2 Targeted Identification of Potential Natural Antiviral Drug Candidates against SARS-CoV-2. Int. J. Biol. Macromol. 2021, 191, 1114–1125.
- Baby, K.; Maity, S.; Mehta, C.H.; Suresh, A.; Nayak, U.Y.; Nayak, Y. SARS-CoV-2 Entry Inhibitors by Dual Targeting TMPRSS2 and ACE2: An in Silico Drug Repurposing Study. Eur. J. Pharmacol. 2021, 896, 173922.
- Telenti, A.; Arvin, A.; Corey, L.; Corti, D.; Diamond, M.S.; García-Sastre, A.; Garry, R.F.; Holmes, E.C.; Pang, P.S.; Virgin, H.W. After the Pandemic: Perspectives on the Future Trajectory of COVID-19. Nature 2021, 596, 495–504.
- Wan, Y.; Shang, J.; Graham, R.; Baric, R.S.; Li, F.; Gallagher, T. Receptor Recognition by the Novel Coronavirus from Wuhan: An Analysis Based on Decade-Long Structural Studies of SARS Coronavirus. J. Virol. 2020, 94, e00127-20.
- Gasmi, A.; Peana, M.; Noor, S.; Lysiuk, R.; Menzel, A.; Gasmi Benahmed, A.; Bjørklund, G. Chloroquine and Hydroxychloroquine in the Treatment of COVID-19: The Never-Ending Story. Appl. Microbiol. Biotechnol. 2021, 105, 1333–1343.
- Kiew, L.-V.; Chang, C.-Y.; Huang, S.-Y.; Wang, P.-W.; Heh, C.-H.; Liu, C.-T.; Cheng, C.-H.; Lu, Y.-X.; Chen, Y.-C.; Huang, Y.-X.; et al. Development of Flexible Electrochemical Impedance Spectroscopy-Based Biosensing Platform for Rapid Screening of SARS-CoV-2 Inhibitors. Biosens. Bioelectron. 2021, 183, 113213.
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural Basis for the Recognition of SARS-CoV-2 by Full-Length Human ACE2. Science 2020, 367, 1444–1448.
- Hamming, I.; Cooper, M.E.; Haagmans, B.L.; Hooper, N.M.; Korstanje, R.; Osterhaus, A.; Timens, W.; Turner, A.J.; Navis, G.; van Goor, H. The Emerging Role of ACE2 in Physiology and Disease. J. Pathol. 2007, 212, 1–11.
- Zhang, H.; Penninger, J.M.; Li, Y.; Zhong, N.; Slutsky, A.S. Angiotensin-Converting Enzyme 2 (ACE2) as a SARS-CoV-2 Receptor: Molecular Mechanisms and Potential Therapeutic Target. Intensive Care Med. 2020, 46, 586–590.
- Hu, Y.; Liu, L.; Lu, X. Regulation of Angiotensin-Converting Enzyme 2: A Potential Target to Prevent COVID-19? Front. Endocrinol. 2021, 12, 1284.
- Patel, V.B.; Zhong, J.-C.; Grant, M.B.; Oudit, G.Y. Role of the ACE2/Angiotensin 1–7 Axis of the Renin–Angiotensin System in Heart Failure. Circ. Res. 2016, 118, 1313–1326.
- Gheblawi, M.; Wang, K.; Viveiros, A.; Nguyen, Q.; Zhong, J.-C.; Turner, A.J.; Raizada, M.K.; Grant, M.B.; Oudit, G.Y. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System. Circ. Res. 2020, 126, 1456–1474.
- Gross, L.Z.F.; Sacerdoti, M.; Piiper, A.; Zeuzem, S.; Leroux, A.E.; Biondi, R.M. ACE2, the Receptor That Enables Infection by SARS-CoV-2: Biochemistry, Structure, Allostery and Evaluation of the Potential Development of ACE2 Modulators. ChemMedChem 2020, 15, 1682–1690.
- Lu, J.; Sun, P.D. High Affinity Binding of SARS-CoV-2 Spike Protein Enhances ACE2 Carboxypeptidase Activity. J. Biol. Chem. 2020, 295, 18579–18588.
- Kiseleva, A.A.; Troisi, E.M.; Hensley, S.E.; Kohli, R.M.; Epstein, J.A. SARS-CoV-2 Spike Protein Binding Selectively Accelerates Substrate-Specific Catalytic Activity of ACE2. J. Biochem. 2021, 170, 299–306.
- Xiao, T.; Lu, J.; Zhang, J.; Johnson, R.I.; McKay, L.G.A.; Storm, N.; Lavine, C.L.; Peng, H.; Cai, Y.; Rits-Volloch, S.; et al. A Trimeric Human Angiotensin-Converting Enzyme 2 as an Anti-SARS-CoV-2 Agent. Nat. Struct. Mol. Biol. 2021, 28, 202–209.
- Jiang, S.; Hillyer, C.; Du, L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359.
- Ji, T.; Liu, Z.; Wang, G.; Guo, X.; Akbar khan, S.; Lai, C.; Chen, H.; Huang, S.; Xia, S.; Chen, B.; et al. Detection of COVID-19: A Review of the Current Literature and Future Perspectives. Biosens. Bioelectron. 2020, 166, 112455.
- Astuti, I. Ysrafil Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2): An Overview of Viral Structure and Host Response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020, 14, 407–412.
- Etienne, E.E.; Nunna, B.B.; Talukder, N.; Wang, Y.; Lee, E.S. COVID-19 Biomarkers and Advanced Sensing Technologies for Point-of-Care (POC) Diagnosis. Bioengineering 2021, 8, 98.
- Yang, J.; Petitjean, S.J.L.; Koehler, M.; Zhang, Q.; Dumitru, A.C.; Chen, W.; Derclaye, S.; Vincent, S.P.; Soumillion, P.; Alsteens, D. Molecular Interaction and Inhibition of SARS-CoV-2 Binding to the ACE2 Receptor. Nat. Commun. 2020, 11, 4541.
- Jackson, C.B.; Farzan, M.; Chen, B.; Choe, H. Mechanisms of SARS-CoV-2 Entry into Cells. Nat. Rev. Mol. Cell Biol. 2021, 23, 3–20.
- Peng, R.; Wu, L.-A.; Wang, Q.; Qi, J.; Gao, G.F. Cell Entry by SARS-CoV-2. Trends Biochem. Sci. 2021, 46, 848–860.
- Udugama, B.; Kadhiresan, P.; Kozlowski, H.N.; Malekjahani, A.; Osborne, M.; Li, V.Y.C.; Chen, H.; Mubareka, S.; Gubbay, J.B.; Chan, W.C.W. Diagnosing COVID-19: The Disease and Tools for Detection. ACS Nano 2020, 14, 3822–3835.
- Parihar, A.; Ranjan, P.; Sanghi, S.K.; Srivastava, A.K.; Khan, R. Point-of-Care Biosensor-Based Diagnosis of COVID-19 Holds Promise to Combat Current and Future Pandemics. ACS Appl. Bio Mater. 2020, 3, 7326–7343.
- Bhalla, N.; Pan, Y.; Yang, Z.; Payam, A.F. Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19. ACS Nano 2020, 14, 7783–7807.
- Zhao, Z.; Huang, C.; Huang, Z.; Lin, F.; He, Q.; Tao, D.; Jaffrezic-Renault, N.; Guo, Z. Advancements in Electrochemical Biosensing for Respiratory Virus Detection: A Review. TrAC Trends Anal. Chem. 2021, 139, 116253.
- Gul, I.; Le, W.; Jie, Z.; Ruiqin, F.; Bilal, M.; Tang, L. Recent Advances on Engineered Enzyme-Conjugated Biosensing Modalities and Devices for Halogenated Compounds. TrAC Trends Anal. Chem. 2021, 134, 116145.
- Rodríguez-Delgado, M.M.; Alemán-Nava, G.S.; Rodríguez-Delgado, J.M.; Dieck-Assad, G.; Martínez-Chapa, S.O.; Barceló, D.; Parra, R. Laccase-Based Biosensors for Detection of Phenolic Compounds. TrAC Trends Anal. Chem. 2015, 74, 21–45.
- Sanches, P.R.S.; Charlie-Silva, I.; Braz, H.L.B.; Bittar, C.; Freitas Calmon, M.; Rahal, P.; Cilli, E.M. Recent Advances in SARS-CoV-2 Spike Protein and RBD Mutations Comparison between New Variants Alpha (B.1.1.7, United Kingdom), Beta (B.1.351, South Africa), Gamma (P.1, Brazil) and Delta (B.1.617.2, India). J. Virus Erad. 2021, 7, 100054.
- Park, S.; Kim, H.; Woo, K.; Kim, J.-M.; Jo, H.-J.; Jeong, Y.; Lee, K.H. SARS-CoV-2 Variant Screening Using a Virus-Receptor-Based Electrical Biosensor. Nano Lett. 2022, 22, 50–57.
- Lee, J.-H.; Lee, Y.; Lee, S.K.; Kim, J.; Lee, C.-S.; Kim, N.H.; Kim, H.G. Versatile Role of ACE2-Based Biosensors for Detection of SARS-CoV-2 Variants and Neutralizing Antibodies. Biosens. Bioelectron. 2022, 203, 114034.
- Torres, M.D.T.; de Araujo, W.R.; de Lima, L.F.; Ferreira, A.L.; de la Fuente-Nunez, C. Low-Cost Biosensor for Rapid Detection of SARS-CoV-2 at the Point of Care. Matter 2021, 4, 2403–2416.
- de Lima, L.F.; Ferreira, A.L.; Torres, M.D.T.; de Araujo, W.R.; de la Fuente-Nunez, C. Minute-Scale Detection of SARS-CoV-2 Using a Low-Cost Biosensor Composed of Pencil Graphite Electrodes. Proc. Natl. Acad. Sci. USA 2021, 118, e2106724118.
- Lee, J.-H.; Choi, M.; Jung, Y.; Lee, S.K.; Lee, C.-S.; Kim, J.; Kim, J.; Kim, N.H.; Kim, B.-T.; Kim, H.G. A Novel Rapid Detection for SARS-CoV-2 Spike 1 Antigens Using Human Angiotensin Converting Enzyme 2 (ACE2). Biosens. Bioelectron. 2021, 171, 112715.
- Yang, Y.; Peng, Y.; Lin, C.; Long, L.; Hu, J.; He, J.; Zeng, H.; Huang, Z.; Li, Z.-Y.; Tanemura, M.; et al. Human ACE2-Functionalized Gold “Virus-Trap” Nanostructures for Accurate Capture of SARS-CoV-2 and Single-Virus SERS Detection. Nano-Micro Lett. 2021, 13, 109.
- Mavrikou, S.; Moschopoulou, G.; Tsekouras, V.; Kintzios, S. Development of a Portable, Ultra-Rapid and Ultra-Sensitive Cell-Based Biosensor for the Direct Detection of the SARS-CoV-2 S1 Spike Protein Antigen. Sensors 2020, 20, 3121.
- Mujica, M.L.; Tamborelli, A.; Castellaro, A.; Barcudi, D.; Rubianes, M.D.; Rodríguez, M.C.; Saka, H.A.; Bocco, J.L.; Dalmasso, P.R.; Rivas, G.A. Impedimetric and Amperometric Genosensors for the Highly Sensitive Quantification of SARS-CoV-2 Nucleic Acid Using an Avidin-Functionalized Multi-Walled Carbon Nanotubes Biocapture Platform. Biosens. Bioelectron. X 2022, 12, 100222.
- Abrego-Martinez, J.C.; Jafari, M.; Chergui, S.; Pavel, C.; Che, D.; Siaj, M. Aptamer-Based Electrochemical Biosensor for Rapid Detection of SARS-CoV-2: Nanoscale Electrode-Aptamer-SARS-CoV-2 Imaging by Photo-Induced Force Microscopy. Biosens. Bioelectron. 2022, 195, 113595.
- Kumar, N.; Shetti, N.P.; Jagannath, S.; Aminabhavi, T.M. Electrochemical Sensors for the Detection of SARS-CoV-2 Virus. Chem. Eng. J. 2022, 430, 132966.
- Luong, A.D.; Buzid, A.; Vashist, S.K.; Luong, J.H.T. Perspectives on Electrochemical Biosensing of COVID-19. Curr. Opin. Electrochem. 2021, 30, 100794.
- Yakoh, A.; Pimpitak, U.; Rengpipat, S.; Hirankarn, N.; Chailapakul, O.; Chaiyo, S. Paper-Based Electrochemical Biosensor for Diagnosing COVID-19: Detection of SARS-CoV-2 Antibodies and Antigen. Biosens. Bioelectron. 2021, 176, 112912.
- Grieshaber, D.; MacKenzie, R.; Vörös, J.; Reimhult, E. Electrochemical Biosensors—Sensor Principles and Architectures. Sensors 2008, 8, 1400–1458.
- Ghazizadeh, E.; Moosavifard, S.E.; Daneshmand, N.; Kaverlavani, S. kamari Impediometric Electrochemical Sensor Based on The Inspiration of Carnation Italian Ringspot Virus Structure to Detect an Attommolar of MiR. Sci. Rep. 2020, 10, 9645.
- Warner, F.J.; Smith, A.I.; Hooper, N.M.; Turner, A.J. What’s New in the Renin-Angiotensin System? Cell. Mol. Life Sci. 2004, 61, 2704–2713.
- Cai, Q.J.; Chan-Park, M.B.; Zhou, Q.; Lu, Z.S.; Li, C.M.; Ong, B.S. Self-Assembled Monolayers Mediated Charge Injection for High Performance Bottom-Contact Poly(3,3′′′-Didodecylquaterthiophene) Thin-Film Transistors. Org. Electron. 2008, 9, 936–943.
- Vezza, V.J.; Butterworth, A.; Lasserre, P.; Blair, E.O.; MacDonald, A.; Hannah, S.; Rinaldi, C.; Hoskisson, P.A.; Ward, A.C.; Longmuir, A.; et al. An Electrochemical SARS-CoV-2 Biosensor Inspired by Glucose Test Strip Manufacturing Processes. Chem. Commun. 2021, 57, 3704–3707.
- Suh, J.-S.; Kim, H.-S.; Kim, T.-J. Development of a SARS-CoV-2-Derived Receptor-Binding Domain-Based ACE2 Biosensor. Sens. Actuators B Chem. 2021, 334, 129663.
- Ferreira, A.L.; de Lima, L.F.; Torres, M.D.T.; de Araujo, W.R.; de la Fuente-Nunez, C. Low-Cost Optodiagnostic for Minute-Time Scale Detection of SARS-CoV-2. ACS Nano 2021, 15, 17453–17462.
- Liu, H.; Dai, E.; Xiao, R.; Zhou, Z.; Zhang, M.; Bai, Z.; Shao, Y.; Qi, K.; Tu, J.; Wang, C.; et al. Development of a SERS-Based Lateral Flow Immunoassay for Rapid and Ultra-Sensitive Detection of Anti-SARS-CoV-2 IgM/IgG in Clinical Samples. Sens. Actuators B Chem. 2021, 329, 129196.
- Moitra, P.; Alafeef, M.; Dighe, K.; Frieman, M.B.; Pan, D. Selective Naked-Eye Detection of SARS-CoV-2 Mediated by N Gene Targeted Antisense Oligonucleotide Capped Plasmonic Nanoparticles. ACS Nano 2020, 14, 7617–7627.
- Ahmed, W.; Angel, N.; Edson, J.; Bibby, K.; Bivins, A.; O’Brien, J.W.; Choi, P.M.; Kitajima, M.; Simpson, S.L.; Li, J.; et al. First Confirmed Detection of SARS-CoV-2 in Untreated Wastewater in Australia: A Proof of Concept for the Wastewater Surveillance of COVID-19 in the Community. Sci. Total Environ. 2020, 728, 138764.
- Corpuz, M.V.A.; Buonerba, A.; Vigliotta, G.; Zarra, T.; Ballesteros, F.; Campiglia, P.; Belgiorno, V.; Korshin, G.; Naddeo, V. Viruses in Wastewater: Occurrence, Abundance and Detection Methods. Sci. Total Environ. 2020, 745, 140910.
- Barcelo, D. An Environmental and Health Perspective for COVID-19 Outbreak: Meteorology and Air Quality Influence, Sewage Epidemiology Indicator, Hospitals Disinfection, Drug Therapies and Recommendations. J. Environ. Chem. Eng. 2020, 8, 104006.
- Medema, G.; Heijnen, L.; Elsinga, G.; Italiaander, R.; Brouwer, A. Presence of SARS-Coronavirus-2 RNA in Sewage and Correlation with Reported COVID-19 Prevalence in the Early Stage of the Epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020, 7, 511–516.
- Langer, J.; de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.J.; Boisen, A.; Brolo, A.G.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117.
- Peng, Y.; Lin, C.; Long, L.; Masaki, T.; Tang, M.; Yang, L.; Liu, J.; Huang, Z.; Li, Z.; Luo, X.; et al. Charge-Transfer Resonance and Electromagnetic Enhancement Synergistically Enabling MXenes with Excellent SERS Sensitivity for SARS-CoV-2 S Protein Detection. Nano-Micro Lett. 2021, 13, 52.
- Yang, L.; Peng, Y.; Yang, Y.; Liu, J.; Huang, H.; Yu, B.; Zhao, J.; Lu, Y.; Huang, Z.; Li, Z.; et al. A Novel Ultra-Sensitive Semiconductor SERS Substrate Boosted by the Coupled Resonance Effect. Adv. Sci. 2019, 6, 1900310.
- Zhang, Y.; Zhen, Y.-R.; Neumann, O.; Day, J.K.; Nordlander, P.; Halas, N.J. Coherent Anti-Stokes Raman Scattering with Single-Molecule Sensitivity Using a Plasmonic Fano Resonance. Nat. Commun. 2014, 5, 4424.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
704
Revisions:
2 times
(View History)
Update Date:
24 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




