
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ashmika Foolchand | -- | 2452 | 2022-11-22 14:25:17 | | | |
| 2 | Lindsay Dong | Meta information modification | 2452 | 2022-11-23 02:18:09 | | |
Video Upload Options
The highly transmittable and infectious COVID-19 remains a major threat worldwide, with the elderly and comorbid individuals being the most vulnerable. While vaccines are currently available, therapeutic drugs will help ease the viral outbreak and prevent serious health outcomes. Epigenetic modifications regulate gene expression through changes in chromatin structure and have been linked to viral pathophysiology. Since epigenetic modifications contribute to the life cycle of the virus and host immune responses to infection, epigenetic drugs are promising treatment targets to ameliorate COVID-19. Deficiency of the multifunctional secosteroid hormone vitamin D is a global health threat. Vitamin D and its receptor function to regulate genes involved in immunity, apoptosis, proliferation, differentiation, and inflammation. Amassed evidence also indicates the biological relations of vitamin D with reduced disease risk, while its receptor can be modulated by epigenetic mechanisms. The immunomodulatory effects of vitamin D suggest a role for vitamin D as a COVID-19 therapeutic agent.
1. Introduction
2. Epigenetics and COVID-19 Infection
2.1. DNA Methylation
2.2. Histone Modifications
2.3. miRNAs
3. Vitamin D Regulation
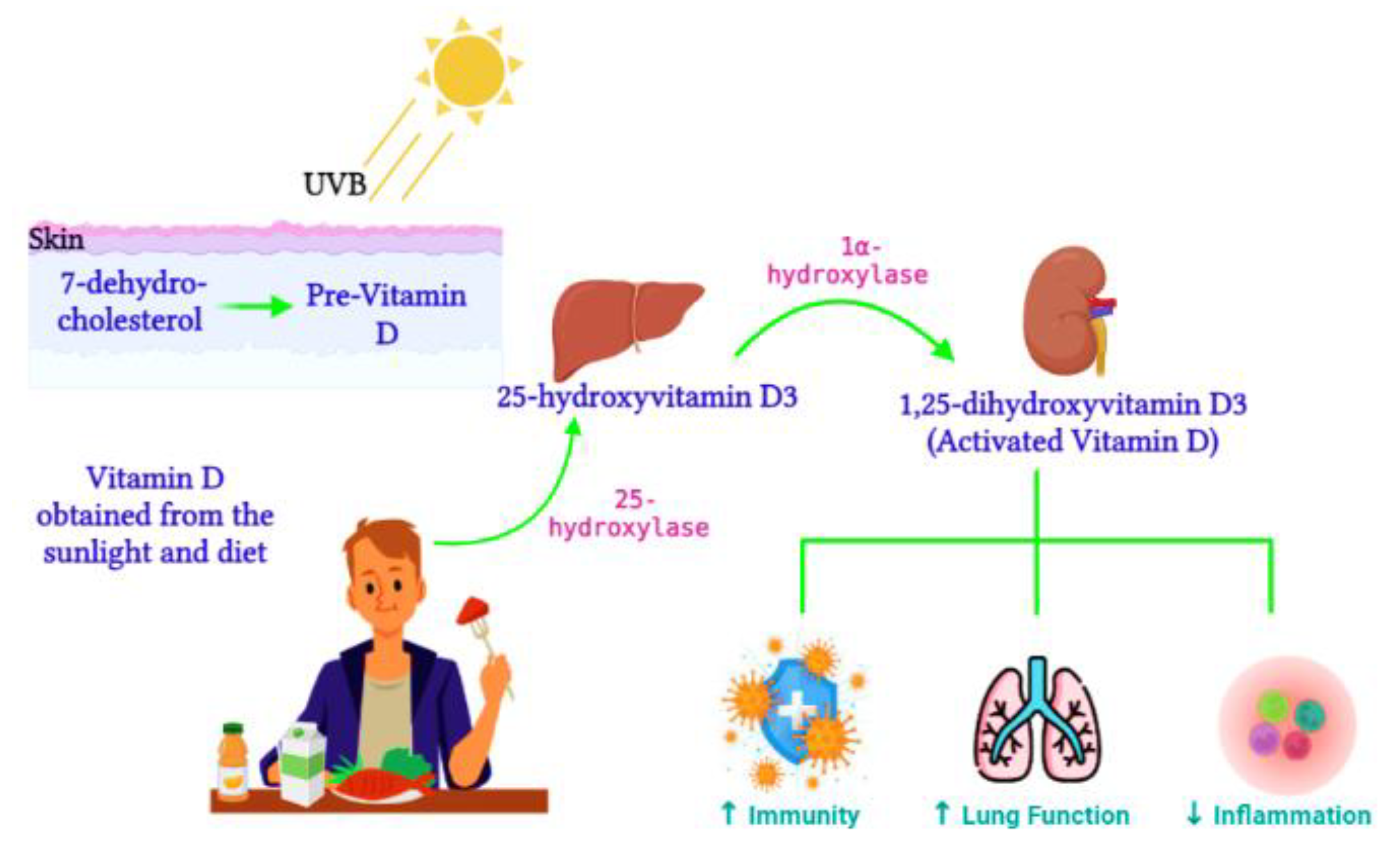
Active vitamin D, 1,25-dihydroxyvitamin D3, is synthesized from the 25-hydroxyvitamin D3 precursor via the 1α-hydroxylase enzyme (CYP27B1) (Figure 1). CYP27B1 is expressed in epithelia which forms the main barrier between the body and its environment. Upon invading pathogens, epithelia respond via its own innate immune system, activating macrophages and dendritic cells to recruit T cells and neutrophils to the infection site [29]. Therefore, during vitamin D insufficiency, reduced availability of the vitamin D precursor would impair active vitamin D production and subsequently alter the innate immune function [30]. Over time, vitamin D deficiency has surged worldwide [31] and has been connected to impaired lung function, enhanced inflammation, and poor immunity [32]. Ligand binding stimulates the translocation of the VDR from the cytosol to the nucleus, where the VDR heterodimerizes with the retinoid X receptor (RXR) to form the VDR/RXR complex. This complex attaches to specific genomic sequences on the promoter region of target genes, known as vitamin D response elements, thus activating or suppressing gene transcription on promoters by recruiting transcription factors and co-regulatory molecules [33]. Active vitamin D is vital for immune regulation. In contrast, vitamin D deficiency was previously associated with the pathogenesis of chronic lung diseases such as asthma and chronic obstructive pulmonary disease in populations exposed to airborne particulates [34]. Vitamin D enhances innate immunity via the antiviral peptide secretion [35], which enhances mucosal defenses. Dietary vitamin D controls genes involved in cell apoptosis, proliferation, differentiation, immune responses, and inflammation [36].
3.1. The Impact of Vitamin D on Epigenetics and Gene Expression
3.2. The Role of Vitamin D in COVID-19 Infections
References
- Yan, R.; Zhang, Y.; Li, Y.; Xia, L.; Guo, Y.; Zhou, Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020, 367, 1444–1448.
- Letko, M.; Marzi, A.; Munster, V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nat. Microbiol. 2020, 5, 562–569.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.H.; Nitsche, A.; et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell 2020, 181, 271–280.e278.
- Sen, R.; Garbati, M.; Bryant, K.; Lu, Y. Epigenetic mechanisms influencing COVID-19. Genome 2021, 64, 372–385.
- Chlamydas, S.; Papavassiliou, A.G.; Piperi, C. Epigenetic mechanisms regulating COVID-19 infection. Epigenetics 2021, 16, 263–270.
- Holmes, L., Jr.; Lim, A.; Comeaux, C.R.; Dabney, K.W.; Okundaye, O. DNA Methylation of Candidate Genes (ACE II, IFN-γ, AGTR 1, CKG, ADD1, SCNN1B and TLR2) in Essential Hypertension: A Systematic Review and Quantitative Evidence Synthesis. Int. J. Environ. Res. Public Health 2019, 16, 4829.
- Bouillon, R.; Carmeliet, G.; Verlinden, L.; van Etten, E.; Verstuyf, A.; Luderer, H.F.; Lieben, L.; Mathieu, C.; Demay, M. Vitamin D and human health: Lessons from vitamin D receptor null mice. Endocr. Rev. 2008, 29, 726–776.
- Azrielant, S.; Shoenfeld, Y. Vitamin D and the Immune System. Isr. Med. Assoc. J. 2017, 19, 510–511.
- Baeke, F.; Takiishi, T.; Korf, H.; Gysemans, C.; Mathieu, C. Vitamin D: Modulator of the immune system. Curr. Opin. Pharmacol. 2010, 10, 482–496.
- Greiller, C.L.; Martineau, A.R. Modulation of the immune response to respiratory viruses by vitamin D. Nutrients 2015, 7, 4240–4270.
- Daniel, C.; Sartory, N.A.; Zahn, N.; Radeke, H.H.; Stein, J.M. Immune Modulatory Treatment of Trinitrobenzene Sulfonic Acid Colitis with Calcitriol Is Associated with a Change of a T Helper (Th) 1/Th17 to a Th2 and Regulatory T Cell Profile. J. Pharmacol. Exp. Ther. 2008, 324, 23.
- Aronson, J.K.; Ferner, R.E. Drugs and the renin-angiotensin system in COVID-19. BMJ 2020, 369, m1313.
- Mehta, P.; McAuley, D.F.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034.
- Dupressoir, A.; Lavialle, C.; Heidmann, T. From ancestral infectious retroviruses to bona fide cellular genes: Role of the captured syncytins in placentation. Placenta 2012, 33, 663–671.
- Obata, Y.; Furusawa, Y.; Hase, K. Epigenetic modifications of the immune system in health and disease. Immunol. Cell. Biol. 2015, 93, 226–232.
- Vavougios, G.D. A data-driven hypothesis on the epigenetic dysregulation of host metabolism by SARS coronaviral infection: Potential implications for the SARS-CoV-2 modus operandi. Med. Hypotheses 2020, 140, 109759.
- Menachery, V.D.; Schäfer, A.; Burnum-Johnson, K.E.; Mitchell, H.D.; Eisfeld, A.J.; Walters, K.B.; Nicora, C.D.; Purvine, S.O.; Casey, C.P.; Monroe, M.E.; et al. MERS-CoV and H5N1 influenza virus antagonize antigen presentation by altering the epigenetic landscape. Proc. Natl. Acad. Sci. USA 2018, 115, E1012–E1021.
- Mueller, A.L.; McNamara, M.S.; Sinclair, D.A. Why does COVID-19 disproportionately affect older people? Aging 2020, 12, 9959–9981.
- Holt, N.R.; Neumann, J.T.; McNeil, J.J.; Cheng, A.C. Implications of COVID-19 for an ageing population. Med. J. Aust. 2020, 213, 342–344.e341.
- Zill, P.; Baghai, T.C.; Schüle, C.; Born, C.; Früstück, C.; Büttner, A.; Eisenmenger, W.; Varallo-Bedarida, G.; Rupprecht, R.; Möller, H.J.; et al. DNA methylation analysis of the angiotensin converting enzyme (ACE) gene in major depression. PLoS ONE 2012, 7, e40479.
- Corley, M.J.; Ndhlovu, L.C. DNA methylation analysis of the COVID-19 host cell receptor, angiotensin I converting enzyme 2 gene (ACE2) in the respiratory system reveal age and gender differences. Preprints 2020, 2020030295.
- Jones, M.J.; Goodman, S.J.; Kobor, M.S. DNA methylation and healthy human aging. Aging Cell 2015, 14, 924–932.
- Pinto, B.G.G.; Oliveira, A.E.R.; Singh, Y.; Jimenez, L.; Gonçalves, A.N.A.; Ogava, R.L.T.; Creighton, R.; Schatzmann Peron, J.P.; Nakaya, H.I. ACE2 Expression Is Increased in the Lungs of Patients With Comorbidities Associated With Severe COVID-19. J. Infect. Dis. 2020, 222, 556–563.
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468.
- de Gonzalo-Calvo, D.; Vea, A.; Bär, C.; Fiedler, J.; Couch, L.S.; Brotons, C.; Llorente-Cortes, V.; Thum, T. Circulating non-coding RNAs in biomarker-guided cardiovascular therapy: A novel tool for personalized medicine? Eur. Heart J. 2019, 40, 1643–1650.
- AbdelHamid, S.G.; Refaat, A.A.; Benjamin, A.M.; Elmawardy, L.A.; Elgendy, L.A.; Manolly, M.M.; Elmaksoud, N.A.; Sherif, N.; Hamdy, N.M. Deciphering epigenetic(s) role in modulating susceptibility to and severity of COVID-19 infection and/or outcome: A systematic rapid review. Environ. Sci. Pollut. Res. Int. 2021, 28, 54209–54221.
- Abedi, F.; Rezaee, R.; Hayes, A.W.; Nasiripour, S.; Karimi, G. MicroRNAs and SARS-CoV-2 life cycle, pathogenesis, and mutations: Biomarkers or therapeutic agents? Cell Cycle 2021, 20, 143–153.
- Farr, R.J.; Rootes, C.L.; Rowntree, L.C.; Nguyen, T.H.O.; Hensen, L.; Kedzierski, L.; Cheng, A.C.; Kedzierska, K.; Au, G.G.; Marsh, G.A.; et al. Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection. PLoS Pathog. 2021, 17, e1009759.
- Bilezikian, J.P.; Bikle, D.; Hewison, M.; Lazaretti-Castro, M.; Formenti, A.M.; Gupta, A.; Madhavan, M.V.; Nair, N.; Babalyan, V.; Hutchings, N. Mechanisms in endocrinology: Vitamin D and COVID-19. Eur. J. Endocrinol. 2020, 183, R133–R147.
- Hewison, M. Vitamin D and the intracrinology of innate immunity. Mol. Cell. Endocrinol. 2010, 321, 103–111.
- Ginde, A.A.; Mansbach, J.M.; Camargo, C.A. Vitamin D, respiratory infections, and asthma. Curr. Allergy Asthma Rep. 2009, 9, 81–87.
- Kumar, T.; Sadoughi, A.; Kohn, N.; Miller, R.; Chandak, T.; Talwar, A. Vitamin D deficiency in advanced lung disease. In A102. Interstitial Lung Disease: Exploring the Pathobiology of Interstitial Lung Disease: What Can We Learn from Histopathology, Genomics, and Biomarkers? American Thoracic Society: New York, NY, USA, 2011; p. A2346.
- Dusso, A.S. Vitamin D receptor: Mechanisms for vitamin D resistance in renal failure. Kidney Int. 2003, 63 (Suppl. 85), S6–S9.
- Herr, C.; Greulich, T.; Koczulla, R.A.; Meyer, S.; Zakharkina, T.; Branscheidt, M.; Eschmann, R.; Bals, R. The role of vitamin D in pulmonary disease: COPD, asthma, infection, and cancer. Respir. Res. 2011, 12, 1–9.
- Wang, T.-T.; Dabbas, B.; Laperriere, D.; Bitton, A.J.; Soualhine, H.; Tavera-Mendoza, L.E.; Dionne, S.; Servant, M.J.; Bitton, A.; Seidman, E.G. Direct and indirect induction by 1, 25-dihydroxyvitamin D3 of the NOD2/CARD15-defensin β2 innate immune pathway defective in Crohn disease. J. Biol. Chem. 2010, 285, 2227–2231.
- Holick, M.F. Vitamin D deficiency. N. Engl. J. Med. 2007, 357, 266–281.
- Meeran, S.M.; Ahmed, A.; Tollefsbol, T.O. Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin. Epigenetics 2010, 1, 101–116.
- Johnson, C.S.; Chung, I.; Trump, D.L. Epigenetic silencing of CYP24 in the tumor microenvironment. J. Steroid. Biochem. Mol. Biol. 2010, 121, 338–342.
- Nishikawa, J.; Kitaura, M.; Matsumoto, M.; Imagawa, M.; Nishihara, T. Difference and similarity of DNA sequence recognized by VDR homodimer and VDR/RXR heterodimer. Nucleic Acids Res. 1994, 22, 2902–2907.
- Fujiki, R.; Kim, M.-s.; Sasaki, Y.; Yoshimura, K.; Kitagawa, H.; Kato, S. Ligand-induced transrepression by VDR through association of WSTF with acetylated histones. EMBO J. 2005, 24, 3881–3894.
- Marik, R.; Fackler, M.; Gabrielson, E.; Zeiger, M.A.; Sukumar, S.; Stearns, V.; Umbricht, C.B. DNA methylation-related vitamin D receptor insensitivity in breast cancer. Cancer Biol. Ther. 2010, 10, 44–53.
- Essa, S.; Denzer, N.; Mahlknecht, U.; Klein, R.; Collnot, E.M.; Tilgen, W.; Reichrath, J. VDR microRNA expression and epigenetic silencing of vitamin D signaling in melanoma cells. J. Steroid. Biochem. Mol. Biol. 2010, 121, 110–113.
- Thorne, J.L.; Maguire, O.; Doig, C.L.; Battaglia, S.; Fehr, L.; Sucheston, L.E.; Heinaniemi, M.; O’Neill, L.P.; McCabe, C.J.; Turner, B.M.; et al. Epigenetic control of a VDR-governed feed-forward loop that regulates p21(waf1/cip1) expression and function in non-malignant prostate cells. Nucleic Acids Res. 2011, 39, 2045–2056.
- Seuter, S.; Pehkonen, P.; Heikkinen, S.; Carlberg, C. Dynamics of 1α,25-dihydroxyvitamin D3-dependent chromatin accessibility of early vitamin D receptor target genes. Biochim. Biophys. Acta 2013, 1829, 1266–1275.
- Pan, L.; Matloob, A.F.; Du, J.; Pan, H.; Dong, Z.; Zhao, J.; Feng, Y.; Zhong, Y.; Huang, B.; Lu, J. Vitamin D stimulates apoptosis in gastric cancer cells in synergy with trichostatin A/sodium butyrate-induced and 5-aza-2′-deoxycytidine-induced PTEN upregulation. FEBS J. 2010, 277, 989–999.
- Abedin, S.A.; Banwell, C.M.; Colston, K.W.; Carlberg, C.; Campbell, M.J. Epigenetic corruption of VDR signalling in malignancy. Anticancer Res. 2006, 26, 2557–2566.
- Hansdottir, S.; Monick, M.M.; Hinde, S.L.; Lovan, N.; Look, D.C.; Hunninghake, G.W. Respiratory epithelial cells convert inactive vitamin D to its active form: Potential effects on host defense. J. Immunol. 2008, 181, 7090–7099.
- Hansdottir, S.; Monick, M.M.; Lovan, N.; Powers, L.; Gerke, A.; Hunninghake, G.W. Vitamin D decreases respiratory syncytial virus induction of NF-κB–linked chemokines and cytokines in airway epithelium while maintaining the antiviral state. J. Immunol. 2010, 184, 965–974.
- Cantorna, M.T.; Snyder, L.; Lin, Y.-D.; Yang, L. Vitamin D and 1, 25 (OH) 2D regulation of T cells. Nutrients 2015, 7, 3011–3021.
- Ahmed, A.; Siman-Tov, G.; Hall, G.; Bhalla, N.; Narayanan, A. Human antimicrobial peptides as therapeutics for viral infections. Viruses 2019, 11, 704.
- Mao, J.; Lin, E.; He, L.; Yu, J.; Tan, P.; Zhou, Y. Autophagy and viral infection. In Autophagy Regulation of Innate Immunity; Springer: Singapore, 2019; pp. 55–78.
- Mushegian, A.A. Autophagy and vitamin D. Sci. Signal. 2017, 10, eaan2526.




