Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | João R. M. Almeida | -- | 1363 | 2022-11-09 13:01:02 | | | |
| 2 | Conner Chen | + 6 word(s) | 1369 | 2022-11-10 03:29:01 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Carneiro, C.V.G.C.; Serra, L.A.; Pacheco, T.F.; Ferreira, L.M.M.; Brandão, L.T.D.; Freitas, M.N.D.M.; Trichez, D.; Almeida, J.R.M.D. Komagataella phaffii Biotechnology. Encyclopedia. Available online: https://encyclopedia.pub/entry/33743 (accessed on 07 February 2026).
Carneiro CVGC, Serra LA, Pacheco TF, Ferreira LMM, Brandão LTD, Freitas MNDM, et al. Komagataella phaffii Biotechnology. Encyclopedia. Available at: https://encyclopedia.pub/entry/33743. Accessed February 07, 2026.
Carneiro, Clara Vida Galrão Corrêa, Luana Assis Serra, Thályta Fraga Pacheco, Letícia Maria Mallmann Ferreira, Lívia Teixeira Duarte Brandão, Mariana Nogueira De Moura Freitas, Débora Trichez, João Ricardo Moreira De Almeida. "Komagataella phaffii Biotechnology" Encyclopedia, https://encyclopedia.pub/entry/33743 (accessed February 07, 2026).
Carneiro, C.V.G.C., Serra, L.A., Pacheco, T.F., Ferreira, L.M.M., Brandão, L.T.D., Freitas, M.N.D.M., Trichez, D., & Almeida, J.R.M.D. (2022, November 09). Komagataella phaffii Biotechnology. In Encyclopedia. https://encyclopedia.pub/entry/33743
Carneiro, Clara Vida Galrão Corrêa, et al. "Komagataella phaffii Biotechnology." Encyclopedia. Web. 09 November, 2022.
Copy Citation
The need for a more sustainable society has prompted the development of bio-based processes to produce fuels, chemicals, and materials in substitution for fossil-based ones. In this context, microorganisms have been employed to convert renewable carbon sources into various products. The methylotrophic yeast Komagataella phaffii has been extensively used in the production of heterologous proteins. The obligate aerobic yeast Komagataella phaffii is a non-pathogenic certified and generally recognized as a safe (GRAS) microorganism. It is classified in the Saccharomycetales order and Saccharomycetaceae family.
Komagataella phaffii
Pichia pastoris
strain engineering
metabolic engineering
1. Introduction
The urgency for effective strategies to mitigate climate change is evident. Environmental issues caused by deliberate petroleum exploitation to produce chemicals, energy, and materials are a major concern regarding global warming. In this context, the global economy needs to be transformed faster, and the circular bioeconomy seems to be the right way to achieve it. This new form of economy aims to reuse, recycle, and remanufacture biomass (from agroecological systems, forestry, and urban wastes) to generate valuable products, such as fuels, biomaterials, and fine chemicals [1]. The circular bioeconomy benefits from integrated frameworks to utilize biomass and address its uses to human needs. One of the major areas that compose this framework is biotechnology, which has the potential to lead bioeconomy global advances even further [2]. The extensive range of roles that biotechnology plays alongside bioengineering provides new techniques to modify the DNA of several microorganisms and plants. Such modifications can improve the utilization of lignocellulosic biomass as raw material through the heterologous protein expression and the production of renewable chemicals, biofuels, and materials [3].
Lignocellulosic biomass polymeric carbohydrates, cellulose, and hemicellulose are mainly composed of glucose and xylose. Glucose is a C6 sugar easily consumed by most microorganisms and can be converted into an extensive range of different chemicals. Xylose, a C5 sugar, can be naturally consumed by filamentous fungi, yeasts, and bacteria. However, this pentose is more challenging to metabolize with the same efficiency as glucose. Nonetheless, xylose can also be converted into various chemicals, such as xylitol [4] and xylonic acid [5]. Recently, advances in genetic manipulation techniques have opened up the range of microorganisms that can produce fine chemicals from glucose and xylose with higher yields and productivity. The yeast Komagataella phaffii is one example of it.
Komagataella phaffii (previously Pichia pastoris) is a well-known methylotrophic yeast. It has many valuable features such as low nutritional requirement, the ability to reach high cell densities (even in acidic culture media), and is widely employed in biotechnological processes to produce heterologous proteins of commercial interest [6]. Therefore, its ability to use methanol as a carbon and energy source, its non-fermentative utilization of glucose under aerobic conditions, and its efficiency to grow on glycerol are some of the main reasons for the preferential choice of this yeast for bioprocess development [7]. These characteristics transformed K. phaffii into a suitable host for many industrial applications. More recently, this yeast has been called the “biotech yeast” for its broad utilization as a cell factory for synthetic biology and metabolic engineering aiming for valuable chemical compound productions (Figure 1) [7][8].
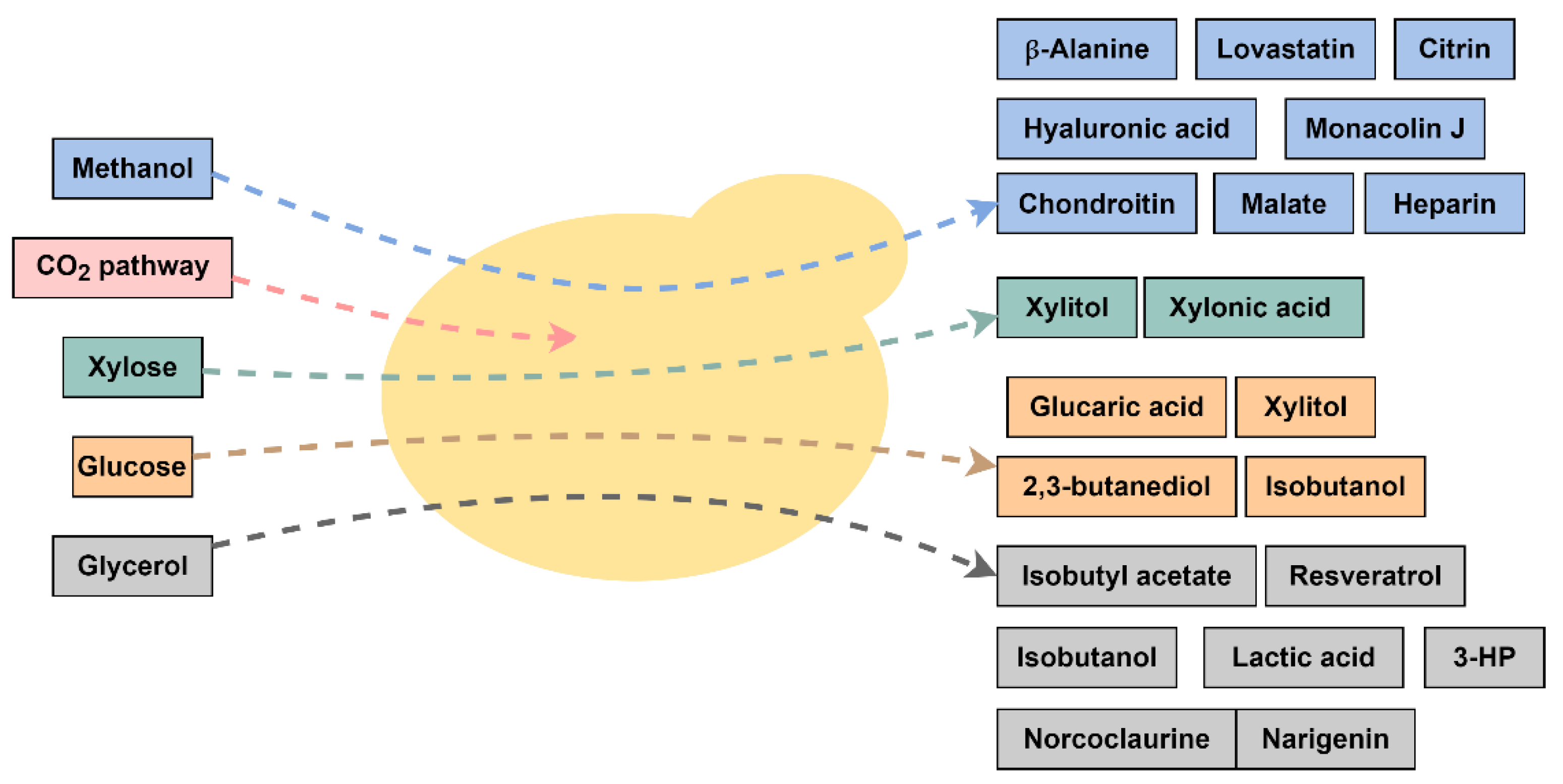
Figure 1. Representative scheme of the different carbon sources and chemicals produced by engineered K. phaffii strains. 3-HP = 3-hydroxy propionic acid.
K. phaffii has already been used as a successful host for heterologous protein production. However, optimizing K. phaffii to produce chemicals is still necessary, mainly aiming at methanol metabolism, fermentation parameters, and the construction of new genetic tools, such as promoters and plasmids. New methods, platforms, and strategies have been developed to enable more feasible ways to engineer this yeast to address these necessities [9]. Strategies to increase the efficiency rate of target integration genes in the K. phaffii genome through the knockout of homologous genes (ku70 [10] and Dnl4p [11]), and also the utilization of CRISPR/Cas9 to facilitate the genome editing of K. phaffii [12], have already been used. Several examples of the available tools to enhance K. phaffii utilization as a good biotechnology host are summarized in [13].
2. Komagataella Taxonomy and Diversity
The obligate aerobic yeast Komagataella phaffii is a non-pathogenic certified and generally recognized as a safe (GRAS) microorganism. It is classified in the Saccharomycetales order and Saccharomycetaceae family. For being capable of using methanol as the only carbon source, K. phaffii is methylotrophic and was anteriorly known as Pichia pastoris. Phylogenetic studies placed P. pastoris in the genus Komagataella proposed by Yamada et al. (1995) after the analysis of partial sequences of rRNAs subunits (18S and 26S) of the 12 strains of methanol assimilating yeasts [14]. Supporting this previous study, Kurtzman and Robnett (1998) compared gene sequences (D1/D2 LSU rRNA) of about 500 species of Ascomycetous yeasts. The multigenic sequence analysis sustained the phylogenetic position of Pichia pastoris in the Komagataella genus [15].
The first species of P. pastoris (described initially as Zygosaccharomyces pastoris) isolated from a chestnut tree was described in 1919 by Guilliermond [16]. Komagataella genus is currently composed of seven species, with most of them (including K. phaffii) isolated from tree exudates in North America and Europe (Table 1) [17]. All species have spherical to ovoid shapes white/cream colonies, and during asexual reproduction, haploid cells multiply via multilateral budding and do not have pseudohyphae or true hyphae. In sexual reproduction (diploid cells), the ascospore is hat-shaped, ranging from 1 to 4 spores (that can be conjugated or not) [18]. The Komagataella species can grow at high cell densities using methanol, glucose, or glycerol as a carbon source with a doubling time of 2 to 3 h, and ammonium can be used as a nitrogen source during growth [17][18]. Recently, it was demonstrated that some species from Komagataella, including K. phaffii, possess the capacity to grow on xylose [19].
Table 1. Komagataella species diversity and its characteristics.
| Type Species | Strain | Genome Size (Mb) | Isolation | Origin | Carbon Sources | Ref |
|---|---|---|---|---|---|---|
| K. pastoris | CBS 704 | 9.6 | Aesculus species | France | Glucose Glycerol Methanol Ethanol Xylose |
ASM170810v1 A |
| K. phaffii | CBS 7435 | 9.4 | Quercus velutina | California, USA | ASM170808v1 A | |
| K. ulmi | CBS 12361 | 9.6 | Ulmus americana | Illinois, USA | [19] | |
| K. kurtzmanii | CBS 12817 | 9.6 | Fir flux | Arizona, USA | ||
| K. mondaviorum | CBS 15017 | 9.5 | Populus deltoides | California, USA | ||
| K. pseudopastoris | CBS 9187 | 10.6 | Salix alba | Hungary | ||
| K. populi | CBS 12362 | 9.3 | Populus deltoides | Illinois, USA |
A: NCBI identifier. Table based on [19].
Since all species have similar phenotypic and fermentative features, they cannot be distinguished using the conventional taxonomy, for example, morphological tests, that are usually used for yeasts. Therefore, gene sequence analysis (D1/D2 LSU rRNA, ITS, EF-1α) and other gene markers tracking are necessary for the correct species placement and identification [18][20]. Several genome sequencing and transcriptome studies have been conducted throughout the years [21][22][23]. A recent study sequenced the genome of all seven type species of Komagataella and its strains (a total of 25 isolates), identifying strains capable of growing using xylose as the sole carbon source, and which have a higher tolerance to stress conditions, such as alkaline pH [19].
The genomes of the two most studied species of the genus, K. pastoris and K. phaffii (previously both species were classified as P. pastoris), have close genome structures to K. ulmi and K. kurtzmanii, respectively. All species are capable of growing in glucose, glycerol, methanol, and ethanol. All 25 strains could grow at acidic pH (4.0) after 7 days, whereas at pH 9.0, all the species were strongly affected. Only the K. pseudopastoris (anteriorly known as P. pseudopastoris) and K. kurtzmanii CBS 12,817 strains were able to adapt and had better growth when compared to the other species [19]. Commonly, non-engineered yeasts in this genus do not present exponential growth on xylose; however, as indicated in the study of Heistinger et al. (2022), all seven species are capable of using this sugar and grow at slow rates. The species K. pastoris CBS 704 and K. populi CBS 12,362 showed the best growth on xylose [19].
Despite the initial studies on Komagataella diversity, the strains of K. phaffii X33 (prototrophic strain) and GS115 (HIS− phenotype) are the ones most used in biotechnological applications, and are mainly used in studies of heterologous expression [20]. In recent years, there have been impressive methodological developments to model and analyze the metabolism of K. phaffii and engineer its genome and metabolic pathways, including improvements in genome modification with homologous recombination and target-guided gene cloning using CRISPR/Cas9 [24]. Indeed, the information about the genetic and physiological profile of this specie and the availability of genetic tools to manipulate it has been crucial for works on metabolic engineering and recombinant protein production studies.
References
- Leong, H.Y.; Chang, C.K.; Khoo, K.S.; Chew, K.W.; Chia, S.R.; Lim, J.W.; Chang, J.S.; Show, P.L. Waste Biorefinery towards a Sustainable Circular Bioeconomy: A Solution to Global Issues. Biotechnol. Biofuels 2021, 14, 1–15.
- Muscat, A.; de Olde, E.M.; Ripoll-Bosch, R.; Van Zanten, H.H.E.; Metze, T.A.P.; Termeer, C.J.A.M.; van Ittersum, M.K.; de Boer, I.J.M. Principles, Drivers and Opportunities of a Circular Bioeconomy. Nat. Food 2021, 2, 561–566.
- Kardung, M.; Cingiz, K.; Costenoble, O.; Delahaye, R.; Heijman, W.; Lovrić, M.; van Leeuwen, M.; M’barek, R.; van Meijl, H.; Piotrowski, S.; et al. Development of the Circular Bioeconomy: Drivers and Indicators. Sustainability 2021, 13, 413.
- Carneiro, C.V.G.; Silva, F.C.D.P.E.; Almeida, J.R. Xylitol Production: Identification and Comparison of New Producing Yeasts. Microorganisms 2019, 7, 484.
- Trichez, D.; Carneiro, C.V.G.C.; Braga, M.; Almeida, J.R.M. Recent Progress in the Microbial Production of Xylonic Acid. World J. Microbiol. Biotechnol. 2022, 38, 127.
- Ata, Ö.; Ergün, B.G.; Fickers, P.; Heistinger, L.; Mattanovich, D.; Rebnegger, C.; Gasser, B. What Makes Komagataella Phaffii Non-Conventional? FEMS Yeast Res. 2021, 21, foab059.
- Bernauer, L.; Radkohl, A.; Lehmayer, L.G.K.; Emmerstorfer-Augustin, A. Komagataella Phaffii as Emerging Model Organism in Fundamental Research. Front. Microbiol. 2021, 11, 1–16.
- Chiang, C.-J.; Ho, Y.-J.; Hu, M.-C.; Chao, Y.-P. Rewiring of Glycerol Metabolism in Escherichia Coli for Effective Production of Recombinant Proteins. Biotechnol. Biofuels 2020, 13, 205.
- Yang, Z.; Zhang, Z. Engineering Strategies for Enhanced Production of Protein and Bio-Products in Pichia Pastoris. Biotechnol. Adv. 2018, 36, 182–195.
- Näätsaari, L.; Mistlberger, B.; Ruth, C.; Hajek, T.; Hartner, F.S.; Glieder, A. Deletion of the Pichia Pastoris KU70 Homologue Facilitates Platform Strain Generation for Gene Expression and Synthetic Biology. PLoS ONE 2012, 7, e39720.
- Ito, Y.; Watanabe, T.; Aikawa, S.; Nishi, T.; Nishiyama, T.; Nakamura, Y.; Hasunuma, T.; Okubo, Y.; Ishii, J.; Kondo, A. Deletion of DNA Ligase IV Homolog Confers Higher Gene Targeting Efficiency on Homologous Recombination in Komagataella Phaffii. FEMS Yeast Res. 2018, 18, foy074.
- Weninger, A.; Hatzl, A.-M.; Schmid, C.; Vogl, T.; Glieder, A. Combinatorial Optimization of CRISPR/Cas9 Expression Enables Precision Genome Engineering in the Methylotrophic Yeast Pichia Pastoris. J. Biotechnol. 2016, 235, 139–149.
- Fischer, J.E.; Glieder, A. Current Advances in Engineering Tools for Pichia Pastoris. Curr. Opin. Biotechnol. 2019, 59, 175–181.
- Yamada, Y.; Matsuda, M.; Maeda, K.; Mikata, K. The Phylogenetic Relationships of Methanol-Assimilating Yeasts Based on the Partial Sequences of 18S and 26S Ribosomal RNAs: The Proposal of Komagataella Gen. Nov. (Saccharomycetaceae). Biosci. Biotechnol. Biochem. 1995, 59, 439–444.
- Kurtzman, C.P.; Robnett, C.J. Identification and Phylogeny of Ascomycetous Yeasts from Analysis of Nuclear Large Subunit (26S) Ribosomal DNA Partial Sequences. Antonie Van Leeuwenhoek 1998, 73, 331–371.
- Guilliermond, A. Zygosaccharomyces Pastori, Nouvelle Espèce de Levures à Copulation Hétérogamique. Bull. Société Mycol. Fr. 1920, 36, 203–211.
- Heistinger, L.; Gasser, B.; Mattanovich, D. Microbe Profile: Komagataella Phaffii: A Methanol Devouring Biotech Yeast Formerly Known as Pichia Pastoris. Microbiology 2020, 166, 614–616.
- Kurtzman, C.P.; Fell, J.W.; Boekhout, T. The Yeasts—A Taxonomy Study; Elsevier Science: Amsterdam, The Netherlands, 2011; Volume 5, ISBN 978-0-08-093127-2.
- Heistinger, L.; Dohm, J.C.; Paes, B.G.; Koizar, D.; Troyer, C.; Ata, Ö.; Steininger-Mairinger, T.; Mattanovich, D. Genotypic and Phenotypic Diversity among Komagataella Species Reveals a Hidden Pathway for Xylose Utilization. Microb. Cell Factories 2022, 21, 70.
- Kurtzman, C.P. Description of Komagataella Phaffii Sp. Nov. and the Transfer of Pichia Pseudopastoris to the Methylotrophic Yeast Genus Komagataella. Int. J. Syst. Evol. Microbiol. 2005, 55, 973–976.
- De Schutter, K.; Lin, Y.-C.; Tiels, P.; Van Hecke, A.; Glinka, S.; Weber-Lehmann, J.; Rouzé, P.; Van de Peer, Y.; Callewaert, N. Genome Sequence of the Recombinant Protein Production Host Pichia Pastoris. Nat. Biotechnol. 2009, 27, 561–566.
- Love, K.R.; Shah, K.A.; Whittaker, C.A.; Wu, J.; Bartlett, M.C.; Ma, D.; Leeson, R.L.; Priest, M.; Borowsky, J.; Young, S.K.; et al. Comparative Genomics and Transcriptomics of Pichia Pastoris. BMC Genom. 2016, 17, 550.
- Mattanovich, D.; Graf, A.; Stadlmann, J.; Dragosits, M.; Redl, A.; Maurer, M.; Kleinheinz, M.; Sauer, M.; Altmann, F.; Gasser, B. Genome, Secretome and Glucose Transport Highlight Unique Features of the Protein Production Host Pichia Pastoris. Microb. Cell Factories 2009, 8, 29.
- Peña, D.A.; Gasser, B.; Zanghellini, J.; Steiger, M.G.; Mattanovich, D. Metabolic Engineering of Pichia Pastoris. Metab. Eng. 2018, 50, 2–15.
More
Information
Subjects:
Biotechnology & Applied Microbiology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.3K
Revisions:
2 times
(View History)
Update Date:
10 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




