
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Eloina Corradi | + 1448 word(s) | 1448 | 2020-11-25 07:20:38 |
Video Upload Options
During neuronal circuit formation, axons progressively develop into a presynaptic compartment aided by extracellular signals. Axons display a remarkably high degree of autonomy supported in part by a local translation machinery that permits the subcellular production of proteins required for their development. MicroRNAs (miRNAs) are critical regulators of this machinery, orchestrating the spatiotemporal regulation of local translation in response to cues. On one hand, a cue-induced relief of miRNA-mediated inhibition leads to bursts of protein translation, on the other hand, a cue-induced miRNA activation, results in reduced protein production. Overall, miRNAs are key elements of the local translation regulatory network controlling axon development.
1.Introduction
During neuronal circuit assembly, the distal tip of the growing axon navigates a complex environment and ultimately reaches its target where it branches and establishes connections with the synaptic partners. Each of these phases of axon development is largely controlled by extracellular signals that induce local cytoskeletal remodeling. As the axon elongates, its tip becomes distant from the cell body and can no longer rely on the soma to promptly supply the proteins that it needs for its development. To overcome this challenge, the axon progressively acquires a high degree of autonomy that enables it to independently manufacture its building blocks. Part of this autonomy is conferred by the delocalization of the translational machinery to the axon to fuel their development through the localized production of proteins.
The molecular mechanisms of mRNA translation in axon development have been clarified through decades of investigation[1][2][3]. Initial studies have documented that selected guidance cues trigger the translation of specific axonal mRNAs within minutes, providing insight into the temporal regulation of mRNA translation and leading to the concept of local translation on demand[4][5][6][7][8]. The advent of large-scale transcriptomics, translatomics, and proteomics analyses have revealed a hitherto unsuspected rich, complex and stage-specific population of axonal transcripts[9][10]. It was confirmed that specific transcripts are selected for translation and that unique protein signatures are synthesized within axons in response to extracellular cues and, unsuspectedly, under basal conditions[11][12]. Moreover, intra-axonal translational events have been observed in adult neurons [11][13]. Thus, beyond the initial evidence of the important role of local translation during neuronal development, such a mechanism appears to be important throughout the neuron’s lifespan, including adulthood [14][15]. Additionally, aspects of the mechanisms underlying the spatial restriction of mRNA translation are starting to be clarified. While it was known that mRNA translation is triggered asymmetrically within the growth cone in relation to the side of cue exposure[5][6], recent studies revealed additional focal hotspots of de novo protein synthesis appear at sites of branch emergence, on RAB7A endosomes, and in association with mitochondria[16][17][18]. Studies from the past 30 years concur to show that the axonal translation machinery triggers the production of the right protein, at the right time, and at the right place to promote axon development. This begs the question of how this process is so exquisitely regulated.
The molecular nature of regulators of axonal mRNA translation and their mechanisms of action are beginning to be elucidated. In general, trans regulators of mRNA translation fall within the two broad categories of RNA binding proteins (RBPs) and non-coding RNAs (ncRNAs), the latter category being overwhelmingly constituted by miRNAs. MiRNAs constitute a large category of short, non-coding RNAs that play widespread regulatory roles in post-transcriptional steps of gene expression, mainly affecting mRNA stability and translation[19]. They act essentially as specificity determinants for effector proteins, which are represented by Argonautes (AGOs) in eukaryotes. MiRNAs act through imperfect matches to their target sequences[20][21], and thereby constitute in vertebrates a vast level of gene expression regulation. In principle, miRNAs are capable of affecting virtually all transcripts within the cell. The scope of miRNA-mediated post-transcriptional gene regulation makes them particularly suited for the spatiotemporal and selective regulation of mRNA translation within axons.
The hunt for regulators of axonal translation has been increasingly zeroing in on the role of miRNAs. Research performed mostly in the last decade has produced a seizable body of evidence showing that miRNAs significantly contribute to support the functional autonomy of the developing axons by exquisitely regulating mRNA translation locally into newly synthesized proteins.
2. Role of miRNAs in Developing Axons
A developing axon displays a large degree of autonomy and relies in part on local mRNA translation to promote its growth, steering, targeting, branching, and synaptogenesis, which are all critical steps in neuronal circuit development. Local translation is a process that is highly regulated in space and time, and axonal miRNAs are emerging as critical components of the mRNA regulatory network. The miRNA repertoire in axons is complex [21][22][23][24][25][26][27][28][29], and initial studies point to an endosome-based mechanism of transport underlying the proper translocation of these small RNAs to the axonal domains where they exert their function[29] . Our current understanding of miRNA-mediated mechanisms in neural circuit assembly points to two key regulatory pathways likely to work in parallel. On the one hand, a cue-induced mRNA de-repression via miRNA inactivation triggers a burst of translation of specific transcripts[28][30][31][32][33]. On the other hand, a cue-induced mRNA repression via NG-miRNA activation induces translation inhibition[29]. Together, the two mechanisms would contribute to regulate growth cone behavior in response to external signals by fine tuning in space and time the translation of mRNAs to ultimately remodel the cytoskeleton (Figure 6). This RNA-based pathway may act alongside the classic protein-based regulatory pathway to confer a higher degree of regulatory potential that supports the precision required in neural circuit development.
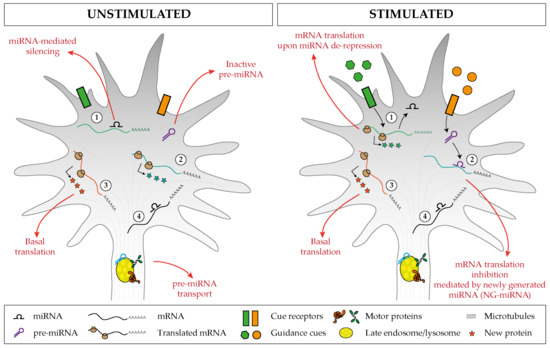
Figure 6. Proposed model of miRNA-mediated regulation of basal and cue-induced translation. (1) Specific miRNAs keep a subset of mRNAs in an untranslated state, but upon stimulation, the miRNA-mediated silencing is relieved, and this induces a burst of mRNA translation. (2) Inactive pre-miRNAs are transported and stored within the axonal compartment (left), and upon cue exposure (right), distinct pre-miRNAs are locally processed and the associated newly generated miRNAs (NG-miRNAs) inhibit the basal translation of given transcripts. Regardless of the stimulus status, (3) specific transcripts undergo basal translation and (4) others are maintained in an untranslated silent state.
Research on the local roles of miRNAs in axons has been intense in the last 10 years, yet many areas remain unexplored. It is still largely unclear how miRNAs are specifically targeted to the axonal compartment and within the axon subregions. Does a zip code exist on such short molecules similar to mRNAs, as shown for miR-29b for nuclear import [34]? Are miRNAs mostly transported as pre-miRNA and processed locally? How widespread is endosome-mediated transport as a mode of transport for miRNAs and RNAs in general? Since the endosome acts as local translational hubs in axons[17], is this organelle crucial in miRNA-regulated translational events in response to external stimuli? Could it spatially restrict the access of newly generated miRNAs to a few targets? Numerous studies have revealed that miRNAs activity is modulated by extrinsic signals. Yet, it is unclear how miRNAs are activated or inactivated upon cue exposure. miRNA inactivation may occur through target-mediated degradation in neurons[35][36]. However two studies have shown that this is not the case, since miRNA levels are not altered [37][38]. MiRNA inactivation could be triggered by miRNA modification[39] or displacement by a molecule competing for the same motif such as an RBP. Circular RNAs (circRNAs) could also be involved, as they can act as miRNA sponges[40][41][42], and it is possible to envision that their levels are increased upon stimulation, leading to the sponging/inactivation of miRNAs. We have uncovered a novel mechanism whereby miRNA activation relies on the local processing of the miRNA precursor[29]. However, other mechanisms may exist.
Clearly, our understanding of miRNA-mediated mechanisms in axon development during circuit assembly remains partial, and further investigations are needed to attain a comprehensive description of how these small non-coding RNAs specifically exert their function. Research in this area will not only help dissecting important molecular mechanisms occurring in neuronal development, but it is assured to also impact research in axon regeneration and in neurological pathologies. Indeed, axon regeneration can be considered a recapitulation of developmental growth. Axonal translation is a crucial mechanism to promote regeneration[43][44], and not surprisingly, miRNAs act as regulators in the regenerative response of injured axons both in the peripheral and central nervous system[45]. In addition, miRNA dysregulation has been linked with neurodevelopmental and neurodegenerative diseases[46]. However, local roles of miRNA in axons in these contexts are largely unexplored. All in all, axonal miRNAs appear to be key regulators throughout the brain lifespan, with potential implication in regenerative processes and in pathological conditions. This characteristic makes them prime targets for therapeutic intervention, and improving our understanding of miRNA-mediated mechanisms might potentially help with treating neuronal trauma and nerve injury in addition to neurodevelopmental and neurodegenerative disorders.
References
- Alvarez, J.; Torres, J.C. Slow axoplasmic transport: A fiction? J. Theor. Biol. 1985, 112, 627–651.
- Alvarez, J. Maintenance of the axoplasm: Can neurones accord with the accepted notions? Neurosci. Lett. 1992, 144, 1–3.
- Alvarez, J.; Giuditta, A.; Koenig, E. Protein synthesis in axons and terminals: Significance for maintenance, plasticity and regulation of phenotype: With a critique of slow transport theory. Prog. Neurobiol. 2000, 62, 1–62.
- Wu, K.Y.; Hengst, U.; Cox, L.J.; Macosko, E.Z.; Jeromin, A.; Urquhart, E.R.; Jaffrey, S.R. Local translation of RhoA regulates growth cone collapse. Nature 2005, 436, 1020–1024.
- Leung, K.-M.; van Horck, F.P.; Lin, A.C.; Allison, R.; Standart, N.; Holt, C.E. Asymmetrical β-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat. Neurosci. 2006, 9, 1247–1256.
- Yao, J.; Sasaki, Y.; Wen, Z.; Bassell, G.J.; Zheng, J.Q. An essential role for beta-actin mRNA localization and translation in Ca2+-dependent growth cone guidance. Nat. Neurosci. 2006, 9, 1265–1273.
- Piper, M.; Anderson, R.; Dwivedy, A.; Weinl, C.; van Horck, F.; Leung, K.M.; Cogill, E.; Holt, C. Signaling mechanisms underlying Slit2-induced collapse of Xenopus retinal growth cones. Neuron 2006, 49, 215–228.
- Cox, L.J.; Hengst, U.; Gurskaya, N.G.; Lukyanov, K.A.; Jaffrey, S.R. Intra-axonal translation and retrograde trafficking of CREB promotes neuronal survival. Nat. Cell Biol. 2008, 10, 149–159.
- Zivraj, K.H.; Tung, Y.C.L.; Piper, M.; Gumy, L.; Fawcett, J.W.; Yeo, G.S.H.; Holt, C.E. Subcellular profiling reveals distinct and developmentally regulated repertoire of growth cone mRNAs. J. Neurosci. 2010, 30, 15464–15478.
- Andreassi, C.; Zimmermann, C.; Mitter, R.; Fusco, S.; De Vita, S.; Devita, S.; Saiardi, A.; Riccio, A. An NGF-responsive element targets myo-inositol monophosphatase-1 mRNA to sympathetic neuron axons. Nat. Neurosci. 2010, 13, 291–301.
- Shigeoka, T.; Jung, H.; Jung, J.; Turner-Bridger, B.; Ohk, J.; Lin, J.Q.; Amieux, P.S.; Holt, C.E. Dynamic Axonal Translation in Developing and Mature Visual Circuits. Cell 2016, 166, 181–192.
- Cagnetta, R.; Frese, C.K.; Shigeoka, T.; Krijgsveld, J.; Holt, C.E. Rapid Cue-Specific Remodeling of the Nascent Axonal Proteome. Neuron 2018, 99, 29–46.e4.
- Hafner, A.-S.; Donlin-Asp, P.G.; Leitch, B.; Herzog, E.; Schuman, E.M. Local protein synthesis is a ubiquitous feature of neuronal pre- and postsynaptic compartments. Science 2019, 364, eaau3644.
- Spaulding, E.L.; Burgess, R.W. Accumulating Evidence for Axonal Translation in Neuronal Homeostasis. Front. Neurosci. 2017, 11, 312.
- Kim, E.; Jung, H. Local mRNA translation in long-term maintenance of axon health and function. Curr. Opin. Neurobiol. 2020, 63, 15–22.
- Wong, H.H.-W.; Lin, J.Q.; Ströhl, F.; Roque, C.G.; Cioni, J.-M.; Cagnetta, R.; Turner-Bridger, B.; Laine, R.F.; Harris, W.A.; Kaminski, C.F.; et al. RNA Docking and Local Translation Regulate Site-Specific Axon Remodeling In Vivo. Neuron 2017, 95, 852–868.e8.
- Cioni, J.-M.; Lin, J.Q.; Holtermann, A.V.; Koppers, M.; Jakobs, M.A.H.; Azizi, A.; Turner-Bridger, B.; Shigeoka, T.; Franze, K.; Harris, W.A.; et al. Late Endosomes Act as mRNA Translation Platforms and Sustain Mitochondria in Axons. Cell 2019, 176, 56–72.e15.
- Spillane, M.; Ketschek, A.; Merianda, T.T.; Twiss, J.L.; Gallo, G. Mitochondria Coordinate Sites of Axon Branching through Localized Intra-Axonal Protein Synthesis. Cell Rep. 2013, 5, 1564–1575.
- Bartel, D.P. MicroRNA Target Recognition and Regulatory Functions. Cell 2009, 136, 215–233.
- Bartel, D.P. Metazoan MicroRNAs. Cell 2018, 173, 20–51.
- Orlangie Natera-Naranjo; Armaz Aschrafi; Anthony E. Gioio; Barry B. Kaplan; Identification and quantitative analyses of microRNAs located in the distal axons of sympathetic neurons. RNA 2010, 16, 1516-1529, 10.1261/rna.1833310.
- Yukio Sasaki; Christina Gross; Lei Xing; Yoshio Goshima; Gary J. Bassell; Identification of axon-enriched MicroRNAs localized to growth cones of cortical neurons. Developmental Neurobiology 2013, 74, 397-406, 10.1002/dneu.22113.
- Melissa L. Hancock; Nicolas Preitner; Jie Quan; John G. Flanagan; MicroRNA-132 Is Enriched in Developing Axons, Locally Regulates Rasa1 mRNA, and Promotes Axon Extension. The Journal of Neuroscience 2013, 34, 66-78, 10.1523/jneurosci.3371-13.2014.
- Nimrod Rotem; Iddo Magen; Ariel Ionescu; Noga Gershoni-Emek; Topaz Altman; Christopher J. Costa; Tal Gradus; Metsada Pasmanik-Chor; Dianna E. Willis; Iddo Z. Ben-Dov; et al.Eran HornsteinEran Perlson ALS Along the Axons – Expression of Coding and Noncoding RNA Differs in Axons of ALS models. Scientific Reports 2017, 7, 44500, 10.1038/srep44500.
- Monichan Phay; Hak Hee Kim; Soonmoon Yoo; Dynamic Change and Target Prediction of Axon-Specific MicroRNAs in Regenerating Sciatic Nerve. PLOS ONE 2015, 10, e0137461, 10.1371/journal.pone.0137461.
- Hak Hee Kim; Paul Kim; Monichan Phay; Soonmoon Yoo; Identification of precursor microRNAs within distal axons of sensory neuron. Journal of Neurochemistry 2015, 134, 193-199, 10.1111/jnc.13140.
- Jose Norberto S. Vargas; Amar N. Kar; Jeffrey A. Kowalak; Jenna Gale; A. Aschrafi; Cai-Yun Chen; Anthony E. Gioio; Barry B. Kaplan; Axonal localization and mitochondrial association of precursor microRNA 338. Cellular and Molecular Life Sciences 2016, 73, 4327-4340, 10.1007/s00018-016-2270-6.
- Anaïs Bellon; Archana Iyer; Simone Bridi; Flora C.Y. Lee; Cesaré Ovando-Vázquez; Eloina Corradi; Sara Longhi; Michela Roccuzzo; Stephanie Strohbuecker; Sindhu Naik; et al.Peter SarkiesEric MiskaCei Abreu-GoodgerChristine E. HoltMarie-Laure Baudet miR-182 Regulates Slit2-Mediated Axon Guidance by Modulating the Local Translation of a Specific mRNA. Cell Reports 2017, 18, 1171-1186, 10.1016/j.celrep.2016.12.093.
- Eloina Corradi; Irene Dalla Costa; Antoneta Gavoci; Archana Iyer; Michela Roccuzzo; Tegan A Otto; Eleonora Oliani; Simone Bridi; Stephanie Strohbuecker; Gabriela Santos‐Rodriguez; et al.Donatella ValdembriGuido SeriniCei Abreu‐GoodgerMarie-Laure Baudet Axonal precursor miRNAs hitchhike on endosomes and locally regulate the development of neural circuits. The EMBO Journal 2020, 39, e102513, 10.15252/embj.2019102513.
- Federico Dajas-Bailador; Boyan Bonev; Patricia Garcez; Peter Stanley; Francois Guillemot; Nancy Papalopulu; microRNA-9 regulates axon extension and branching by targeting Map1b in mouse cortical neurons. Nature Neuroscience 2012, 15, 697-699, 10.1038/nn.3082.
- Bin Wang; Lin Pan; Manyi Wei; Qiong Wang; Wen-Wen Liu; Nuoxin Wang; Xing-Yu Jiang; Xu Zhang; Lan Bao; FMRP-Mediated Axonal Delivery of miR-181d Regulates Axon Elongation by Locally Targeting Map1b and Calm1. Cell Reports 2015, 13, 2794-2807, 10.1016/j.celrep.2015.11.057.
- Tao Yang; Huai Huang; Qiangqiang Shao; Shirley Yee; Tanushree Majumder; Guofa Liu; miR-92 Suppresses Robo1 Translation to Modulate Slit Sensitivity in Commissural Axon Guidance. Cell Reports 2018, 24, 2694-2708.e6, 10.1016/j.celrep.2018.08.021.
- Cristiano Lucci; Raquel Mesquita Ribeiro; Alex Rathbone; Federico A Dajas-Bailador; Spatiotemporal regulation of GSK3β levels by miRNA-26a controls axon development in cortical neurons.. Development 2020, 147, dev180232, 10.1242/dev.180232.
- Hwang, H.-W.; Wentzel, E.A.; Mendell, J.T. A hexanucleotide element directs microRNA nuclear import. Science 2007, 315, 97–100.
- Bitetti, A.; Mallory, A.C.; Golini, E.; Carrieri, C.; Carreño Gutiérrez, H.; Perlas, E.; Pérez-Rico, Y.A.; Tocchini-Valentini, G.P.; Enright, A.J.; Norton, W.H.J.; et al. MicroRNA degradation by a conserved target RNA regulates animal behavior. Nat. Struct. Mol. Biol. 2018, 25, 244–251.
- Kleaveland, B.; Shi, C.Y.; Stefano, J.; Bartel, D.P. A Network of Noncoding Regulatory RNAs Acts in the Mammalian Brain. Cell 2018, 174, 350–362.e17.
- Bellon, A.; Iyer, A.; Bridi, S.; Lee, F.C.Y.; Ovando-Vázquez, C.; Corradi, E.; Longhi, S.; Roccuzzo, M.; Strohbuecker, S.; Naik, S.; et al. miR-182 Regulates Slit2-Mediated Axon Guidance by Modulating the Local Translation of a Specific mRNA. Cell Rep. 2017, 18, 1171–1186.
- Wang, B.; Pan, L.; Wei, M.; Wang, Q.; Liu, W.-W.; Wang, N.; Jiang, X.-Y.; Zhang, X.; Bao, L. FMRP-Mediated Axonal Delivery of miR-181d Regulates Axon Elongation by Locally Targeting Map1b and Calm1. Cell Rep. 2015, 13, 2794–2807.
- Krol, J.; Loedige, I.; Filipowicz, W. The widespread regulation of microRNA biogenesis, function and decay. Nat. Rev. Genet. 2010, 11, 597–610.
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Zheng, Q.; Bao, C.; Guo, W.; Li, S.; Chen, J.; Chen, B.; Luo, Y.; Lyu, D.; Li, Y.; Shi, G.; et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 2016, 7, 11215.
- Zhao, Y.; Alexandrov, P.N.; Jaber, V.; Lukiw, W.J. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7). Genes 2016, 7, 116.
- Twiss, J.L.; Smith, D.S.; Chang, B.; Shooter, E.M. Translational control of ribosomal protein L4 mRNA is required for rapid neurite regeneration. Neurobiol. Dis. 2000, 7, 416–428.
- Terenzio, M.; Koley, S.; Samra, N.; Rishal, I.; Zhao, Q.; Sahoo, P.K.; Urisman, A.; Marvaldi, L.; Oses-Prieto, J.A.; Forester, C.; et al. Locally translated mTOR controls axonal local translation in nerve injury. Science 2018, 359, 1416–1421.
- Roitbak, T. MicroRNAs and Regeneration in Animal Models of CNS Disorders. Neurochem. Res. 2020, 45, 188–203.
- Martino, S.; di Girolamo, I.; Orlacchio, A.; Datti, A.; Orlacchio, A. MicroRNA Implications across Neurodevelopment and Neuropathology. Available online: https://www.hindawi.com/journals/bmri/2009/654346/ (accessed on 5 November 2020).




