Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Ruphi Naz | -- | 2861 | 2022-11-01 09:40:40 | | | |
| 2 | Conner Chen | + 24 word(s) | 2885 | 2022-11-02 02:33:42 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Naz, R.; Khan, A.; Alghamdi, B.S.; Ashraf, G.M.; Alghanmi, M.; Ahmad, A.; Bashir, S.S.; Haq, Q.M.R. Function, Expression and Applications of AtGluR Genes. Encyclopedia. Available online: https://encyclopedia.pub/entry/32285 (accessed on 08 February 2026).
Naz R, Khan A, Alghamdi BS, Ashraf GM, Alghanmi M, Ahmad A, et al. Function, Expression and Applications of AtGluR Genes. Encyclopedia. Available at: https://encyclopedia.pub/entry/32285. Accessed February 08, 2026.
Naz, Ruphi, Andleeb Khan, Badrah S. Alghamdi, Ghulam Md Ashraf, Maimonah Alghanmi, Altaf Ahmad, Sheikh Shanawaz Bashir, Qazi Mohd Rizwanul Haq. "Function, Expression and Applications of AtGluR Genes" Encyclopedia, https://encyclopedia.pub/entry/32285 (accessed February 08, 2026).
Naz, R., Khan, A., Alghamdi, B.S., Ashraf, G.M., Alghanmi, M., Ahmad, A., Bashir, S.S., & Haq, Q.M.R. (2022, November 01). Function, Expression and Applications of AtGluR Genes. In Encyclopedia. https://encyclopedia.pub/entry/32285
Naz, Ruphi, et al. "Function, Expression and Applications of AtGluR Genes." Encyclopedia. Web. 01 November, 2022.
Copy Citation
Arabidopsis thaliana contains 20 glutamate receptor genes (AtGluR) comparable to the human ionotropic glutamate (iGluRs) receptor. Many studies have proved that AtGluR genes are involved in a number of plant growth and physiological activities, such as in the germination of seeds, roots, abiotic and biotic stress, and cell signaling, which clarify the place of these genes in plant biology.
fluorescence resonance energy transfer
glutamate receptors
ligand binding domain
1. Clade I
1.1. AtGluR-1.1
The glutamate receptor genes (AtGluR)-1.1 gene has been reported to play a role in potential activity by regulating and signaling abscisic acid (ABA) biosynthesis in A. thaliana [1]. AtGluR-1.1 also regulates C, N, and water balance, which are essential for plant growth. The GUS expression was first observed in stipules and the collette region of 7-day-old Arabidopsis plants. Later, its expression was found in the leaf margins and in the cells of the lateral roots (Figure 1). A low expression of GluR-1.1 was also detected in the flowers, siliques, and reproductive organs [2].
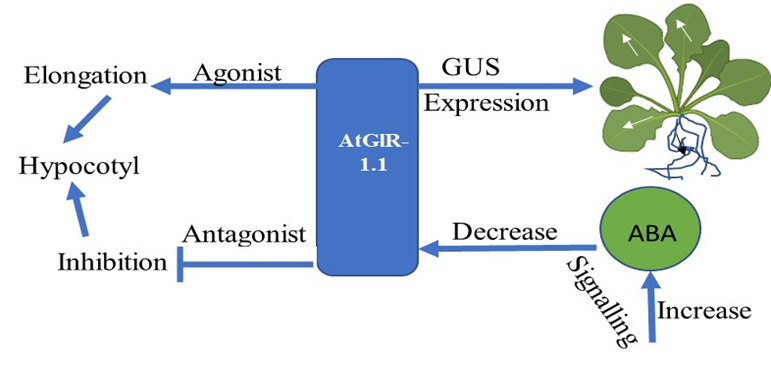
Figure 1. Flowchart showing the expression and function of the AtGluR-1.1 gene.
1.2. AtGluR-1.2 and 1.3
Zheng et al. [3] demonstrated that GluR-1.2 and -1.3 regulate cold tolerance in Arabidopsis owing to cold stress by stimulating endogenous jasmonate synthesis, and these genes also play a crucial role in the downstream CBF/DREB1pathway during cold stress [3][4] (Figure 2). A fluorescent tag was ligated with GluR-1.2 and -1.3 to detect its expression, and an increased fluorescence was observed in the plasma membrane of Nicotiana benthamiana [3].
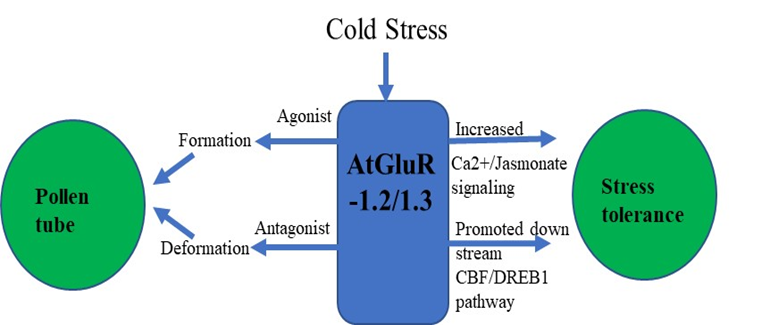
Figure 2. Flowchart of AtGluR-1.2/-1.3.
1.3. AtGluR-1.4
The expression of GFP-tagged AtGluR-1.4 in the plasma membrane of wild-type Arabidopsis plants was studied. StREM1.3, a plasma membrane marker tagged with red fluorescent protein (RFP), was also employed for co-localization [5]. However, Roy et al. [6] detected a varying expression of AtGluR-1.4 in different cells of the same plant, as well as in distinct plants. In addition, Ca2+ depolarization in leaf cells is perhaps caused by amino acid signaling [5] (Figure 3).
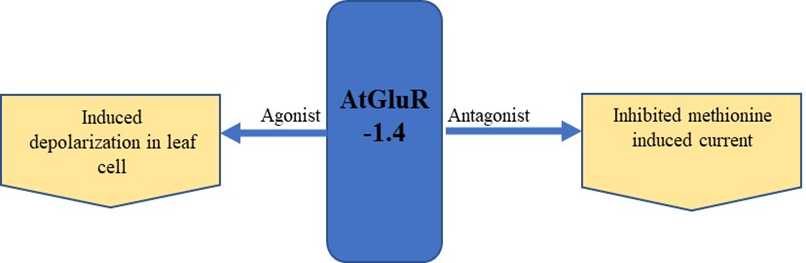
Figure 3. Expression and effect of the agonist and antagonist on AtGluR-1.4.
2. Clade II
2.1. AtGluR-2.1
On the basis of the phylogenetic and expression analysis of the glutamate receptor, Chiu et al. [2] reported that expression of a clade-II gene (GluR-2.1) was observed in the shoot of a 5-day-old plant that showed similarity with GluR-1.1. In addition, GluR-2.1 was also detected in the radical of emerging seedling, and it was expended in all cells of the root, except in the root apex of 3-day-old seedling. However, a slight expression was detected in the reproductive organs and no such expression was shown in the siliques or flowers [2]. Roy et al. [6] found that AtGluR-2.1 was expressed in 4-week-old leaf tissue. The pretreatment of glutamate in plants enhanced the upregulation of the gene expression and the accumulation of amino acids [7] (Figure 4).
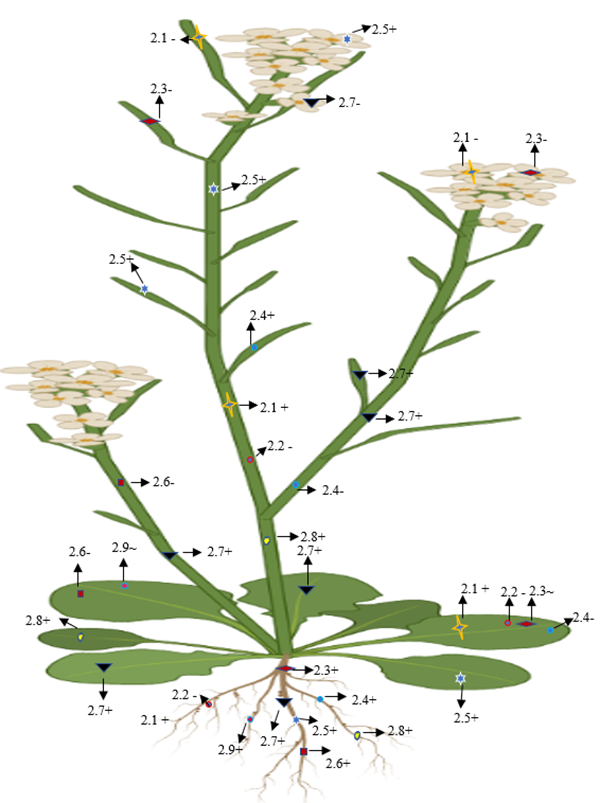
Figure 4. Diagrammatic representation of the presence, absence, and lower expression of Clade II genes in Arabidopsis. + (Expression), (no expression), ~ (low expression).
2.2. AtGluR-2.2
The expression of AtGluR-2.2 was reported in the root, whereas it was lost in reproductive organs and the leaves [2]. A similar observation was reported by Roy et at. [6], where AtGluR-2.2 was expressed in the root but was absent in leaf, stem, and petiole part of the plant.
2.3. AtGluR-2.3
The expression of AtGluR-2.3 was quite different, as it was restricted in 8-week-old plants; after that, it was seen in the root tissue [2]. AtGluR-2.3 was not expressed in the flowers and silique, while a deficient expression was detected in the leaves of the developing plant [6].
2.4. AtGluR-2.4
In addition, AtGluR-2.4 was expressed in the root and silique, but the expression was completely stopped in stem, petioles, and leaf tissues [2][6][8].
2.5. AtGluR-2.5
Based on the mRNA expression analysis, Chiu et al. [2] reported that AtGluR-2.5 was expressed in the whole plant tissues.
2.6. AtGluR-2.6
2.7. AtGluR-2.7
2.8. AtGluR-2.8
Along with the root and shoot, a good expression was also observed in the leaf mesophyll cells around the vascular bundles with a GUS straining and expression became higher in the leaves during the senescence stage [8][9] (Figure 4).
2.9. AtGluR-2.9
Similar to the other Clade II receptors, the AtGluR-2.9 gene was also expressed in the root [8]; however, Roy et al. [6] observed that gene 2.9 either showed a lower expression in the leaves or was completely stopped (Figure 4).
3. Clade III
3.1. AtGluR-3.1
In order to comprehend the expression and function of AtGluR-3.1, Kong et al. [10] investigated AtGluR-3.1 in depth and fused a green fluorescent protein (GFP) tag with GluR-3.1 and 3.5, determining that these proteins were situated in the plasma membrane and were expressed in the guard cells. Furthermore, the study found that these receptors were expressed in the seedling cells, other than the guard cells. Cho et al. [11], on the other hand, suggested that an increased expression of AtGluR-3.1 was seen in the guard cells, followed by the mesophyll cells. Furthermore, the deregulated expression of AtGluR-3.1 altered the stomatal closure, without impairing cytosolic Ca2+.
3.2. AtGluR-3.2
The expression of AtGluR-3.2 was confirmed in all parts of the plant and was highly expressed in the root cells [12]. The exogenous supply of Ca2+ demonstrated the function of AtGluR-3.2 to overcome the Ca2+ deprived condition, which was induced through overexpressed AtGluR-3.2 in transgenic plants [13].
3.3. AtGluR-3.3
According to Li et al. [14], supplementation of exogenous GSH activates the AtGluR-3.3 and participates in the early transcriptional process of the leaves. However, the genetic process underlying GSH and the independent expression of the gene is not known. Furthermore, root gravitropism in AtGluR-3.3 mutant lines is likely to diminish amino acid regulated Ca2+ signaling [15].
3.4. AtGluR-3.4
Among the other AtGluRs, AtGluR-3.4 is strongly expressed in guard cells, vascular bundles, roots, and mesophyll and stem. AtGluR-3.4 responds under abiotic stimuli such as cold and touch in a Ca2+ dependent manner. Along with the different expression, a weak expression was also observed in the cortex, root epidermis, and hairs. However, the activity of the cold expression was halted by adding lanthanum (Ca2+ channel blocker). Moreover, a mutation in AtGluR-3.4 enhanced sensitivity towards ABA, which caused an impact on seed germination [16].
3.5. AtGluR-3.5
The expression of AtGluR-3.5 predominantly appeared in Arabidopsis germinating seeds, where this receptor enhances the Ca2+ concentration. No significant expression was seen in the dry and mature seeds. However, repression in AtGluR-3.5 impacted Ca2+ signaling and was more sensitive to ABA, which exhibited a delay in seed germination. In contrast, a higher expression of AtGluR-3.5 was less sensitive to ABA and led to early germination. The site of expression of AtGluR-3.5 was confirmed by creating a transgenic plant with the GUS gene and it was identified that AtGluR-3.5 was expressed in the cotyledons of the germinating embryo [17].
3.6. AtGluR-3.6
The study of AtGluR-3.6 demonstrated that the expression of AtGluR-3.6 is based in the developmental stages of the roots. Singh et al. [18] reported a higher expression level in the early stages of root tissues than the mature root tissues. The overexpression of AtGluR-3.6 promoted root growth. Furthermore, KRP4 (kip related protein) played a crucial role in maintaining AtGluR-3.1-related root meristem [18]. Cell cycle regulation also influenced the formation of the main and lateral roots [18][19]. Furthermore, it has been suggested that AtGluR-rec-3.6 may play a role in leaf wound signaling via the jasmonate pathway [20]. Other research indicated that AtGluR-3.6/3.3 played a vital role to induce Ca2+ elevation on the aphid feeding sites [21].
3.7. AtGluR-3.7
Considerably, research on AtGluR-3.7 has found that this receptor is likely expressed in every plant cell and serves an important function as an ion transporter in Arabidopsis [6]. Further research has demonstrated that AtGluR-3.7 responds to salt stress in Arabidopsis via calcium signaling and is involved in seed germination and regulation [22][23]. The overexpression of AtGluR-3.7 provoked a marked increase in root growth.
4. AtGluRs Gene Respond to Environmental Stress
4.1. Stress Due to Salt
Salt stress alters plant behavior and adaption mechanisms, which are usually mediated by the Ca2+ signaling network [24][25][26][27][28]. The study of Cheng et al. [29] demonstrated that AtGluR-3.7 played a significant role during seed germination under salt stress conditions. Furthermore, Cheng et al. [30] confirmed that salt stress treatment on Arabidopsis wild and mutant plants revealed that AtGluR-(3.4-1/2) of mutants were more sensitive in seed germination when compared with the wild type. An expansion study showed NaCl caused a rise in Ca2+ wave in the wild variety of plants, which was blocked by inserting an animal glutamate receptor antagonist (DNQX (6,7-dinitroquinoxaline-2,3-dione)). However, an increase in Ca2+, triggered by NaCl, was impaired in mutated AtGluRs (3.4-1/2). In addition, more Na+ was accumulated in the mutant during salt stress than in the wild-type seeds. Overexpressed AtGluR-3.4 was highly resistant to abscisic acid [30]. Furthermore, Wang et al. [22] reported in their study that AtGluR-3.7, in conjunction with other proteins (14-3-3), plays a potential role in Arabidopsis under salt stress by influencing the Ca2+ signaling pathway. As a result of these findings, seed germination under salt conditions was regulated by Ca2+ inflow, which was modulated by AtGluRs-3.4/3.7. Therefore, the Clade III gene, GluR-3.7, plays a significant role in salt stress in A. thaliana [31].
4.2. Stress Due to Cold
In addition to salt stress, cold stress is also another factor that harms plants. In 1999, Thomashow [4] reported that temperate region originating plants such as spinach and Arabidopsis showed cold tolerance upon exposure to low temperatures. This phenomenon is known as cold acclimation. In addition to the plasma membrane, chloroplast also plays a sensing role at low temperatures. Earlier, Meyerhoff et al. [24] suggested that because of the osmotic stress in Arabidopsis, AtGluR-3.4 was expressed in an ABA-independent fashion and provides strength in fast signaling via Ca2+ ion.
In transgenic plants, cold and glutamate (Glu) were able to induce Ca2+ ion channels, which was blocked by the antagonist (DNQX/6-cyano-7-nitroquinoxaline-2,3-dione (CNQX)). The study of Hu et al. [32] indicated that during cold stress, phytohormones altered their activity and response towards plants. Further research revealed that the membrane protein sensed cold stress and perhaps activated Ca2+ signal transduction pathways. These signals may lead to activating the downstream transcriptional regulatory cascade and allow them to cope and survive in cold stress [33][34][35][36][37]. Thus, exposure to cold temperature plants induces tolerance in cold conditions, as well as a massive alteration in gene expression and Ca2+ signal transduction pathways. In addition, Zheng et al. [3] unveiled that AtGluRs-1.2 and -1.3 mutants performed a positive role towards cold stress in Arabidopsis, whereas with the influence of the plant hormone jasmonate, the activity of AtGluRs-1.2/-1.3 mutants to cold stress was reduced. Furthermore, under a freezing environment, the expression of these mutants was lower in the C-repeat binding factor/DRE binding factor 1 (CBF/DREB1) transcriptional regulatory pathway compared with the wild-type plant. In brief, Zheng et al. [3] suggested that AtGluRs-1.2/1.3 could increase cold tolerance by trigging endogenous jasmonate accumulation (Figure 5).
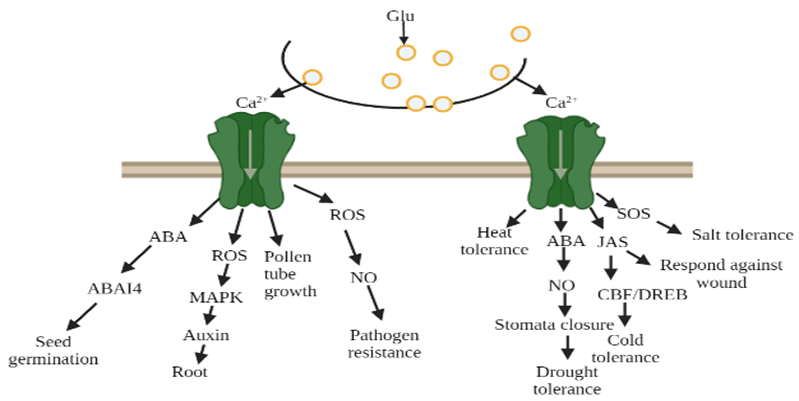
Figure 5. Glutamate receptors mediated by Ca2+ ion due to environmental stress, glutamate (Glu), abscisic acid (ABA), reactive oxygen species (ROS), nitric oxide (NO), jasmonate (Jas).
4.3. Stress Due to Drought
Drought stress is a severe environmental barrier to plant productivity. Drought stress impacts different physiological processes and reduces crop yield [38][39]. Water scarcity has been observed at a morphological to molecular level in plants. Drought stress severely impacts on the photosynthetic abilities of plants, which imbalances metabolic activities. When faced with stress, plants modify their strategies to allow them to be capable of resisting and adapting to drought conditions [38][39]. After many debates on the closure of the stomata, its impact on drought stress or metabolic impairment was found. It has been proven that stomatal closure is one of the main factors for controlling water deficits as well as gas exchange during stress, which is regulated by the co-ordination of different signaling molecules, including Ca2+, H2S, reactive oxygen species, ABA, and nitric oxide NO [7][38][39]. The study of Yoshida et al. [39] demonstrated that glutamate (Glu), a signaling molecule that increases Ca2+ in guard cells, is responsible for closing the stomata and preventing light from opening up the stomata in Arabidopsis, as well as in Vicia faba. Furthermore, to discover the complete relevance of glutamate receptors with stomatal closure due to glutamate, different Ca2+ chelators such as EGTA (extracellular Ca2+ chelator) and BAPTA-AM (intracellular chelator) were used, and it was found that these chelators prevent the stomata from being closed, which is induced by glutamate, and show that the influx of Ca2+ into the cytosol is required. They also observed that Glu-dependent Ca2+ flow triggers Ca2+ dependent protein kinase and endorse the SLAC activity to close the stomata [39]. Other than Arabidopsis, Glu treatment in Brassica responds by triggering Ca2+ signaling, which increases proline accumulation due to drought stress and manages drought tolerance [40]. In addition, Philippe et al. [41] used animal glutamate receptor antagonist-AP-5, DNQX, in Medicago truncatula and reported that these antagonists were responsible for reducing NO accumulation, which showed the least effect on stomata closure. Furthermore, two glutamate receptors obtained from rice (GLR1 and GLR2) exhibited drought tolerance in Arabidopsis [42].
5. Role of AtGluRs in Plant Defense Signaling
Plants have defended themselves by evolving various defense tactics against the opponents, which take the shape of insects, pests, and other herbivores. Plants use both chemical and mechanical means to detect herbivores. Many recent studies have focused on monitoring early defense signaling to protect them from herbivore attacks in the form of fluorescent-based genetically encoded nanosensors to detect in real time signaling and modulate the ion channels that are commonly associated with plant−herbivore interactions [43]. Furthermore, a number of studies have shown that Glu receptors protect plants from insect injury. Because of insect activity on plants, Glu receptors increase Ca2+ flow, causing defensive signals to change in order for the plants to survive [44][45].
In an experiment when A. thaliana was wounded by S. littoralis, it was found that AtGluR-3.3 induced Ca2+ ion flow to reduce the surface potential caused by the larvae and showed a potential role in altering the defense signaling in plants [20]. Glu receptors are involved in plant defense signaling as they are activated by the wound [19][20]. In addition, recent research suggested that GluRs-3.3/3.6 modulates Ca2+ ion flux to defend the plants against wound-creating organisms [43][46]. Initially, the research of Kang et al. [47] showed that in transgenic Arabidopsis, overexpressed radish GluR intermingles with the increased expression of some defense-linked genes and accelerates the resistance against fungal pathogens. Afterwards, pharmacological studies showed that ionotropic glutamate receptors (iGluRs) antagonists were found to take part in the immune response in seedlings of A. thaliana [23] and in the suspension culture of tobacco [45]. In AtGluR-3.3 mutants, because of the attack of a bacterial pathogen (Pseudomonas syringae), susceptibility increased with regard to immune response, which exhibited the inadequacy in the activity of defense-related gene expression against infection [14]. Although almost all AtGluR genes take part in the defense mechanism, AtGluR-3.3 was found to be more responsible, as in P syringae, activation of the defense gene in response to enhancing immunity against infection depends on AtGluR-3.3 [48]. A study suggested that the glutamate receptors associated with Ca2+ influx are essential for initiating downstream signaling processes towards the plant defense. Further involvement of AtGluR-3.3 is important for resistance infected by Hyaloperonospora arabidopsis (oomycete pathogen) [49]. It has been found that the Clade III gene fully participates in the defensive mechanism against mechanical wounding caused by feeding insects and pests [20][50].
6. Developed FRET Based Glutamate Receptors Nanosensors
Modern biosensors based on fluorescence resonance energy transfer (FRET) provide a potential tool. FRET has become an advanced phenomenon for monitoring the interactions, such as molecule to molecule or protein to protein, or conformational changes (Figure 6). The biggest advantage is that the distances are obviously shorter than the diffraction limits of other microscopies. Fluorescent proteins (FP) are used as an essential component in the creation of biosensors. This method is based on the excitation of the donor fluorophore, followed by the non-radiative transfer of the excited energy to the related other fluorophore protein, known as the acceptor molecule [51][52]. Various FRET-based nanosensors have been constructed, but the contents here concentrated on FRET-based approaches to glutamate receptors. Glutamate receptors are not only for animal neurons and glial cells [53]. These receptors exist in the entire world, as well as eukaryotes and prokaryotes [54][55][56]. Okumoto et al. [57] were the first to construct a glutamate ratiometric sensor by sandwiching a truncated binding protein (gltl) of glutamate/aspartate between a fluorophore protein pair (enhanced cyan fluorescent protein (ECFP), mVenus). Hires et al. [58] introduced EYFP (enhanced yellow fluorescent protein) in lieu of m-Venus and conducted systematic monitoring of the glutamate affinities. Next, Marvin et al. [59][60] proposed that single-wavelength fluorescence receptors (FR) could detect neurotransmitters with a great spatial and temporal resolution. They created a number of glutamate iGluSnFR sensor versions employing blue, green, cyan, and yellow color emissions and circular permutated cp-GFP, with activities ranging from sub-micromolecular to millimolar.
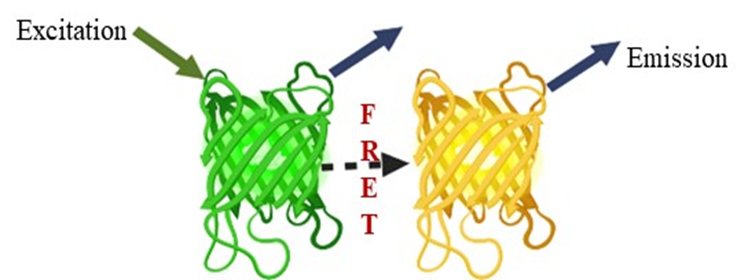
Figure 6. FRET phenomenon, a potential tool.
The plant glutamate receptor protein family emerged as a binding cassette for neurotransmitters with the introduction of animal glutamate nanosensors. In a similar way to how glutamate acts in neurons, it also aids in the flow of impulses in plants [19][61]. Plants use the fluorescent indicator protein for glutamate (FLIPE) [61] (Figure 7). According to Forde and Roberts [54], glutamate receptor proteins have been found to bind glutamate/4-aminobutanoic acid (GABA) and amino acids such as glutamate/glycine. The high-affinity GABA transporter (AtGAT1) in A. thaliana may be required for GABA sensor function, as found by Meyer et al. [62]. To change the spectral range of the glutamate sensor, Wu et al. [63] created circular permutated (cp) R-iGluSnFR1 using red Glu sensing FR and noncircular permutated (ncp) Rncp-iGluSnFR1.
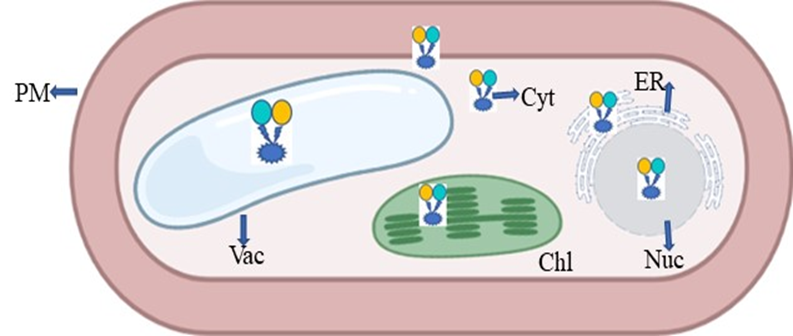
Figure 7. Expression of FRET in the form of nanosensor in a plant cell; PM—plasma membrane; ER—endoplasmic reticulum; Chl—chloroplast; Vac—vacuole; Nuc—nucleus; Cyt—cytoplasm.
References
- Kang, J.; Mehta, S.; Turano, F.J. The putative glutamate receptor 1.1 (AtGlR1.1) in Arabidopsis thaliana regulates abscisic acid biosynthesis and signaling to control development and water loss. Plant Cell Physiol. 2004, 45, 1380–1389.
- Chiu, J.C.; Brenner, E.D.; DeSalle, R.; Nitabach, M.N.; Holmes, T.C.; Coruzzi, G.M. Phylogenetic and expression analysis of the glutamate-receptorlike gene family in Arabidopsis thaliana. Mol. Bio. Evol. 2002, 19, 1066–1082.
- Zheng, Y.; Luo, L.; Wei, J.; Chen, Q.; Yang, Y.; Hu, X.; Kong, X. The glutamate receptors AtGLR1.2 and AtGLR1.3 increase cold tolerance by regulating jasmonate signaling in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2018, 506, 895–900.
- Thomashow, M.F. Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Ann. Rev. Plant Physiol. Plant Mole. Biol. 1999, 50, 571–599.
- Tapken, D.; Anschütz, U.; Liu, L.; Huelsken, T.; Seebohm, G.; Becker, D.; Hollmann, M. A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci. Signal. 2013, 6, ra47.
- Roy, S.J.; Gilliham, M.; Berger, B.; Essah, P.A.; Cheffings, C.; Miller, A.J.; Davenport, R.J.; Liu, L.H.; Skynner, M.J.; Davies, J.M.; et al. Investigating glutamate receptor-like gene co-expression in Arabidopsis thaliana. Plant Cell Environ. 2008, 31, 861–871.
- Qiu, X.M.; Sun, Y.Y.; Ye, X.Y.; Li, Z.G. Signaling role of glutamate in plants. Front. Plant Sci. 2020, 10, 1743.
- Weiland, M.; Mancuso, S.; Baluska, F. Signalling via glutamate and GLRs in Arabidopsis thaliana. Funct. Plant Biol. 2015, 43, 1–25.
- Gilliham, M.; Campbell, M.; Dubos, C.; Becker, D.; Davenport, R. The Arabidopsis thaliana glutamate-like receptor family (AtGLR). In Communication in Plants; Baluška, F., Mancuso, S., Volkmann, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2006; pp. 187–204.
- Kong, D.; Hu, H.C.; Okuma, E.; Lee, Y.; Lee, H.S.; Munemasa, S.; Cho, D.; Ju, C.; Pedoeim, L.; Rodriguez, B.; et al. L-Met Activates Arabidopsis GLR Ca2+ channels upstream of ROS production and regulates stomatal movement. Cell Rep. 2016, 17, 2553–2561.
- Cho, D.; Kim, S.A.; Murata, Y.; Lee, S.; Jae, S.K.; Nam, H.G.; Kwak, J.M. Deregulated expression of the plant glutamate receptor homolog AtGLR3.1 impairs long-term Ca2+ -programmed stomatal closure. Plant J. 2009, 58, 437–449.
- Vincill, E.D.; Bieck, A.M.; Spalding, E.P. Ca2+ conduction by an amino acid-gated ion channel related to glutamate receptors. Plant Physiol. 2012, 159, 40–46.
- Kim, S.A.; Kwak, J.; Jae, S.K.; Wang, M.H.; Nam, H. Overexpression of the AtGluR2 gene encoding an Arabidopsis homolog of mammalian glutamate receptors impairs calcium utilization and sensitivity to ionic stress in transgenic plants. Plant Cell Physiol. 2001, 42, 74–84.
- Li, F.; Wang, J.; Ma, C.; Zhao, Y.; Wang, Y.; Hasi, A.; Qi, Z. Glutamate receptor-like channel3.3 is involved in mediating glutathione-triggered cytosolic calcium transients, transcriptional changes, and innate immunity responses in Arabidopsis. Plant Physiol. 2013, 162, 1497–1509.
- Miller, N.D.; Durham Brooks, T.L.; Assadi, A.H.; Spalding, E. P Detection of a gravitropism phenotype in glutamate receptor-like 3.3 mutants of Arabidopsis thaliana using machine vision and computation. Genetics 2010, 186, 585–593.
- Cheng, Y.; Zhang, X.; Sun, T.; Tian, Q.; Zhang, W.H. Glutamate receptor homolog3.4 is involved in regulation of seed germination under salt stress in Arabidopsis. Plant Cell Physiol. 2018, 59, 978–988.
- Kong, D.; Ju, C.; Parihar, A.; Kim, S.; Cho, D.; Kwak, J.M. Arabidopsis glutamate receptor homolog3.5 modulates cytosolic Ca2+ level to counteract effect of abscisic acid in seed germination. Plant Physiol. 2015, 167, 1630–1642.
- Singh, S.K.; Chien, C.T.; Chang, I.F. The Arabidopsis glutamate receptor-like gene GLR3.6 controls root development by repressing the Kip-related protein gene KRP4. J. Exp. Bot. 2016, 67, 1853–1869.
- Toyota, M.; Spencer, D.; Sawai-Toyota, S.; Jiaqi, W.; Zhang, T.; Koo, A.J.; Howe, G.A.; Gilroy, S. Glutamate triggers long-distance, calcium-based plant defense signaling. Science 2018, 361, 1112–1115.
- Mousavi, S.A.; Chauvin, A.; Pascaud, F.; Kellenberger, S.; Farmer, E.E. Glutamate-receptor like genes mediate leaf-to-leaf wound signalling. Nature 2013, 500, 422–426.
- Vincent, T.R.; Avramova, M.; Canham, J.; Higgins, P.; Bilkey, N.; Mugford, S.T.; Pitino, M.; Toyota, M.; Gilroy, S.; Miller, A.J.; et al. Interplay of Plasma Membrane and Vacuolar Ion Channels, Together with BAK1, Elicits Rapid Cytosolic Calcium Elevations in Arabidopsis during Aphid Feeding. Plant Cell 2017, 29, 1460–1479.
- Wang, P.H.; Lee, C.E.; Lin, Y.S.; Lee, M.H.; Chen, P.Y.; Chang, H.C.; Chang, I.F. The Glutamate Receptor-Like Protein GLR3.7 Interacts With 14-3-3ω and Participates in Salt Stress Response in Arabidopsis thaliana. Front. Plant Sci. 2019, 10, 1169.
- Kwaaitaal, M.; Huisman, R.; Maintz, J.; Reinstädler, A.; Panstruga, R. Ionotropic glutamate receptor (iGluR)-like channels mediate MAMP-induced calcium influx in Arabidopsis thaliana. Biochem. J. 2011, 440, 355–365.
- Meyerhoff, O.; Müller, K.; Roelfsema, M.R.G.; Latz, A.; Lacombe, B.; Hedrich, R.; Dietrich, P.; Becker, D. AtGLR3.4, a glutamate receptor channel-like gene is sensitive to touch and cold. Planta 2005, 222, 418–427.
- Sivaguru, M.; Pike, S.; Gassmann, W.; Baskin, T.I. Aluminum rapidly depolymerizes cortical microtubules and depolarizes the plasma membrane. Evidence that these Responses are Mediated by a Glutamate Receptor. Plant Cell Physiol. 2003, 44, 667–675.
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 19, 371–379.
- Muchate, N.S.; Nikalje, G.C.; Rajurkar, N.S.; Suprasanna, P.; Nikam, T.D. Plant salt stress: Adaptive responses, tolerance mechanism and bioengineering for salt tolerance. Bot. Rev. 2016, 82, 371–406.
- Saddhe, A.A.; Malvankar, M.R.; Karle, S.B.; Kumar, K. Reactive nitrogen species: Paradigms of cellular signaling and regulation of salt stress in plants. Environ. Exp. Bot. 2019, 161, 86–97.
- Cheng, Y.; Tian, Q.; Zhang, W.H. Glutamate receptors are involved in mitigating effects of amino acids on seed germination of Arabidopsis thaliana under salt stress. Environ. Exp. Bot. 2016, 130, 68–78.
- Chen, P.Y.; Hsu, C.Y.; Lee, C.E.; Chang, I.F. Arabidopsis glutamate receptor GLR3.7 is involved in abscisic acid response. Plant Signal. Behav. 2021, 16, 1997513.
- Hu, Y.; Jiang, L.; Wang, F.; Yu, D. Jasmonate regulates the inducer of cbf expression–C-repeat binding factor/dre binding factor1 Cascade and Freezing Tolerance in Arabidopsis. Plant Cell 2013, 25, 2907–2924.
- Ding, Y.; Jia, Y.; Shi, Y.; Zhang, X.; Song, C.; Gong, Z.; Yang, S. OST1-mediated BTF3L phosphorylation positively regulates CBFs during plant cold responses. EMBO J. 2018, 37, e98228.
- Zhu, S.; Stein, R.A.; Yoshioka, C.; Lee, C.H.; Goehring, A.; Mchaourab, H.S.; Gouaux, E. Mechanism of NMDA receptor inhibition and activation. Cell 2016, 165, 704–714.
- Ding, Y.; Shi, Y.; Yang, S. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 2019, 222, 1690–1704.
- Friedrich, T.; Faivre, L.; Bäurle, I.; Schubert, D. Chromatin-based mechanisms of temperature memory in plants. Plant Cell Environ. 2018, 42, 762–770.
- Zhou, A.; Liu, E.; Li, H.; Li, Y.; Feng, S.; Gong, S.; Wang, J. PsCor413pm2, a Plasma membrane-localized, cold-regulated protein from Phlox subulata, confers low temperature tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2579.
- Hasanuzzaman, M.; Alam, M.M.; Nahar, K.; Mohsin, S.M.; Bhuyan, M.B.; Parvin, K.; Hawrylak-Nowak, B.; Fujita, M. Silicon-induced antioxidant defense and methylglyoxal detoxification works coordinately in alleviating nickel toxicity in Oryza sativa L. Ecotoxicology 2019, 28, 261–276.
- Laxa, M.; Liebthal, M.; Telman, W.; Chibani, K.; Dietz, K.J. The role of the plant antioxidant system in drought tolerance. Antioxidants 2019, 8, 94.
- Yoshida, R.; Mori, I.C.; Kamizono, N.; Shichiri, Y.; Shimatani, T.; Miyata, F.; Honda, K.; Iwai, S. Glutamate functions in stomatal closure in Arabidopsis and fava bean. J. Plant Res. 2016, 129, 39–49.
- La, V.H.; Lee, B.R.; Islam, M.T.; Park, S.H.; Jung, H.I.; Bae, D.W.; Kim, T.H. Characterization of salicylic acid-mediated modulation of the drought stress responses: Reactive oxygen species, proline, and redox state in Brassica napus. Environ. Exp. Bot. 2019, 157, 1–10.
- Philippe, F.; Verdu, I.; Morère-Le Paven, M.C.; Limami, A.M.; Planchet, E. Involvement of Medicago truncatula glutamate receptor-like channels in nitric oxide production under short-term water deficit stress. J. Plant Physiol. 2019, 6, 1–6.
- Lu, G.; Wang, X.; Liu, J.; Yu, K.; Gao, Y.; Liu, H.; Wang, C.; Wang, W.; Wang, G.; Liu, M.; et al. Application of T-DNA activation tagging to identify glutamate receptor-like genes that enhance drought tolerance in plants. Plant Cell Rep. 2014, 33, 617–631.
- Gandhi, A.; Kariyat, R.; Harikishore, A.; Ayati, M.; Bhunia, A.; Sahoo, N. Deciphering the Role of Ion Channels in Early Defense Signaling against Herbivorous Insects. Cells 2021, 10, 2219.
- Dennison, K.L.; Spalding, E.P. Glutamate-gated calcium fluxes in Arabidopsis. Plant Physiol. 2000, 124, 1511–1514.
- Vatsa, P.; Chiltz, A.; Bourque, S.; Wendehenne, D.; Garcia-Brugger, A.; Pugin, A. Involvement of putative glutamate receptors in plant defence signaling and NO production. Biochimie 2011, 93, 2095–2101.
- Shao, Q.; Gao, Q.; Lhamo, D.; Zhang, H.; Luan, S. Two glutamate- and pH-regulated Ca2+ channels are required for systemic wound signaling in Arabidopsis. Sci. Signal. 2020, 13, 1453.
- Kang, S.; Kim, H.B.; Lee, H.; Choi, J.Y.; Heu, S.; Oh, C.J.; Kwon, S.I.; An, C.S. Overexpression in Arabidopsis of a plasma membrane-targeting glutamate receptor from small radish increases glutamate-mediated Ca2+ influx and delays fungal infection. Mol. Cells 2006, 21, 418–427.
- Qi, Z.; Stephens, N.R.; Spalding, E.P. Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 2006, 142, 963–971.
- Manzoor, H.; Kelloniemi, J.; Chiltz, A.; Wendehenne, D.; Pugin, A.; Poinssot, B.; Garcia-Brugger, A. Involvement of the glutamate receptor AtGLR3.3 in plant defense signaling and resistance to Hyaloperonospora Arabidopsis. Plant J. 2013, 76, 466–480.
- de Bruxelles, G.L.; Roberts, M.R. Signals regulating multiple responses to wounding and herbivores. Cri. Rev. Plant Sci. 2001, 20, 487–521.
- Li, G.; Kong, W.; Zhao, M.; Lu, S.; Gong, P.; Chen, G.; Xia, L.; Wang, H.; You, J.; Wu, Y. A fluorescence resonance energy transfer (FRET) based “Turn-On” nanofluorescence sensor using a nitrogen-doped carbon dot-hexagonal cobalt oxyhydroxide nanosheet architecture and application to alpha-glucosidase inhibitor screening. Biosens. Bioelectron. 2016, 79, 728–735.
- Leopold, S.J.; Watson, J.A.; Jeeyapant, A.; Simpson, J.A.; Phu, N.H.; Hien, T.T.; Day, N.P.J.; Dondorp, A.M.; White, N.J. Investigating causal pathways in severe falciparum malaria: A pooled retrospective analysis of clinical studies. PLoS Med. 2019, 16, e1002858.
- Harada, K.; Kamiya, T.; Tsuboi, T. Gliotransmitter release from astrocytes: Functional, developmental, and pathological implications in the brain. Front. Neurosci. 2015, 9, 499.
- Forde, B.G.; Roberts, M.R. Glutamate receptor-like channels in plants: A role as amino acid sensors in plants. F1000 Prime Rep. 2014, 6, 37.
- Pin, J.P.; Galvez, T.; Prezeau, L. Evolution, structure, and activation mechanism of family 3/C G-protein-coupled receptors. Pharmacol. Ther. 2003, 98, 325–354.
- Ribeiro, P.; Patocka, N. Neurotransmitter transporters in schistosomes: Structure, function and prospects for drug discovery. Parasitol. Int. 2013, 62, 629–638.
- Okumoto, S.; Looger, L.L.; Micheva, K.D.; Reimer, R.J.; Smith, S.J.; Frommer, W.B. Detection of glutamate release from neurons by genetically encoded surface-displayed FRET nanosensors. Proc. Natl. Acad. Sci. USA 2005, 102, 8740–8745.
- Hires, S.A.; Zhu, Y.; Tsien, R.Y. Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc. Natl. Acad. Sci. USA 2008, 105, 4411–4416.
- Marvin, J.S.; Borghuis, B.G.; Tian, L.; Cichon, J.; Harnett, M.T.; Akerboom, J.; Gordus, A.; Renninger, S.L.; Chen, T.W.; Bargmann, C.I.; et al. An optimized fluorescent probe for visualizing glutamate neurotransmission. Nat. Methods 2013, 10, 162–170.
- Marvin, J.S.; Scholl, B.; Wilson, D.E.; Podgorski, K.; Kazemipour, A.; Müller, J.A.; Schoch, S.; Quiroz, F.J.U.; Rebola, N.; Bao, H.; et al. Stability, affinity, and chromatic variants of the glutamate sensor iGluSnFR. Nat. Methods 2018, 11, 936–939.
- Castro-Rodrı’guez, V.; Kleist, T.J.; Gappel, N.M.; Okumoto, S.; Machado, M.; Denyer, T.; Timmermans, M.C.; Frommer, W.B.; Wudick, M.M. Noxious effects of cell surface display glutamate sensors on plant growth and development. bioRxiv 2020.
- Meyer, A.; Eskandari, S.; Grallath, S.; Rentsch, D. AtGAT1, a high affinity transporter for gamma-aminobutyric acid in Arabidopsis thaliana. J. Biol. Chem. 2006, 281, 7197–7204.
- Wu, J.; Abdelfattah, A.S.; Zhou, H.; Ruangkittisakul, A.; Qian, Y.; Ballanyi, K.; Campbell, R.E. Genetically encoded glutamate indicators with altered color and topology. ACS Chem. Biol. 2018, 13, 1832–1837.
More
Information
Subjects:
Plant Sciences
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
02 Nov 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




