Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Robert Cormier | -- | 3312 | 2022-08-18 17:09:59 | | | |
| 2 | Dean Liu | -40 word(s) | 3272 | 2022-08-19 03:40:04 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Wang, Z.; Cormier, R.T. Golden Syrian Hamster Models for Cancer Research. Encyclopedia. Available online: https://encyclopedia.pub/entry/26288 (accessed on 07 February 2026).
Wang Z, Cormier RT. Golden Syrian Hamster Models for Cancer Research. Encyclopedia. Available at: https://encyclopedia.pub/entry/26288. Accessed February 07, 2026.
Wang, Zhongde, Robert T. Cormier. "Golden Syrian Hamster Models for Cancer Research" Encyclopedia, https://encyclopedia.pub/entry/26288 (accessed February 07, 2026).
Wang, Z., & Cormier, R.T. (2022, August 18). Golden Syrian Hamster Models for Cancer Research. In Encyclopedia. https://encyclopedia.pub/entry/26288
Wang, Zhongde and Robert T. Cormier. "Golden Syrian Hamster Models for Cancer Research." Encyclopedia. Web. 18 August, 2022.
Copy Citation
The golden Syrian hamster (Mesocricetus auratus) has long been a valuable rodent model of human diseases, especially infectious and metabolic diseases. Hamsters have also been valuable models of several chemically induced cancers such as the DMBA-induced oral cheek pouch cancer model.
golden Syrian hamster
cancer
KCNQ1
TP53
IL2RG
1. History of Hamsters as Animal Models in Cancer Research
The golden Syrian hamster (Mesocricetus auratus) has been demonstrated to be especially effective in modeling human disorders such as diet-induced early atherosclerosis, inflammatory myopathies, emerging viral infectious diseases, Clostridium difficile infection, pancreatitis, diet-induced obesity, insulin resistance, and lipid metabolism [1][2][3][4][5][6][7][8]. Notably, for several decades hamsters have also been successfully used to model virally and chemically induced cancers [9][10][11][12][13][14][15][16][17][18][19][20][21]. An excellent example is oral cancers induced via topical administration of carcinogenic chemicals such as 7,12-dimethylbenz[a]anthracene (DMBA). Head and neck cancer is the sixth most common cancer worldwide with oral cancers comprising roughly half of all head and neck cancers, and despite advances in therapy morbidity and mortality for oral cancers remain high. For several decades the hamster model of sequential oral carcinogenesis following topical administration of DMBA to the hamster cheek pouch has been shown to be a superior animal model for recapitulation of human oral cancers, from premalignant to malignant states. The hamster oral cheek model is especially informative for oral cancers that develop from the use of smokeless tobacco products (STPs) that are chewed or dipped in the human cheek pouch. It is noted that there are an estimated 300 million STP users worldwide including 3% of US adults and 6% of US high school students [15]. For the treatment of the growing number of oral cancers, the hamster oral cheek model has also been effective in the development of new therapies such as boron neutron capture therapy (BNCT) [11][15][17][18][19][20]. Another example of a long-used effective chemically induced cancer in the hamster is the N-Nitrosobis(2-oxopropyl)amine (BOP)-treated model of pancreatic cancer [12][16]. Pancreatic cancer is an extremely lethal disease with close to 450,000 deaths each year worldwide and a poor 5-year survival rate of 10%. The hamster BOP model closely resembles the stages in human pancreatic cancer as it causes pancreatic intraepithelial neoplasia (PanIN), a premalignant stage of pancreatic cancer that progresses to severe pancreatic ductal adenocarcinoma as observed in humans, including harboring similar genetic mutations such as in K-ras, CDKN2A, and SMAD4 [16]. Finally, another example of the long-standing use of the hamster in cancer research is its employment as an animal model of oncolytic adenoviruses and to evaluate new antiviral drugs [13].
However, until recently a significant challenge for the use of hamsters in modeling cancer and other human diseases, compared, for example, with mouse models, was the inability to generate genetically engineered hamsters. The first steps to overcome this limitation came with the full sequencing and annotation of the hamster genome and transcriptome [14][21]. Now, this barrier has been surmounted by the group, resulting in gene KOs and knockins in the hamster by employing CRISPR/Cas9-mediated gene targeting, piggyBac-mediated transgenesis, and pronuclear injection [22][23]. Researchers described the phenotypes of the first three genetically engineered hamster cancer models, Kos in the tumor suppressors TP53 and KCNQ1, and a KO in the IL2RG gene, whose first application was to provide a non-murine model to study X-linked severe combined immunodeficiency (XSCID) and the infectious diseases associated with IL2RG deficiency.
2. Genetic Engineering in the Golden Syrian Hamster
Despite the long history of the golden Syrian hamster being used as a laboratory organism for biomedical research, the application of hamsters had been severely limited by the lack of technologies to manipulate the hamster genome. A few years ago, researchers took up the challenge to establish genetic engineering technologies in the hamster and successfully applied the CRISPR/Cas9 technique to conducting gene KO experiments in this species [22][23]. Researchers subsequently also established CRISPR/cas9-mediated gene-knockin (KI) and edited techniques in the hamster. Recently, researchers also developed piggyBac-mediated transgenic techniques in the hamster and produced transgenic hamsters expressing human genes, including the human angiotensin-converting enzyme 2 (hACE2) transgenic hamster lines as a model for COVID-19 [24][25][26]. With these genetic engineering techniques, researchers have produced a long list of the first genetically engineered hamster lines which are being used by many laboratories in several countries to study a variety of aspects of basic biology and human conditions, including cancer, metabolic syndrome, circadian rhythm, and viral infections.
3. KCNQ1, TP53, and IL2RG Genetically Engineered Hamster Cancer Models
3.1. KCNQ1 Knockout Hamster Model
KCNQ1 is a developmentally imprinted potassium channel gene that is widely expressed in both human and rodent tissues. Among other functions, KCNQ1 is well known for its voltage-gated interaction with its heterodimeric partner KCNE1 to regulate cardiac myocyte repolarization. KCNQ1 mutations in humans cause a range of disease pathologies in humans including cardiac arrhythmia, inner ear defects, and gastric hyperplasia. Notably, work by the group has shown that KCNQ1 acts as a tumor suppressor gene in the gastrointestinal tract in both humans and mice [27][28]. For example, the maintenance of the expression of KCNQ1 was associated with a significant survival advantage in human colorectal cancer patients with stage IV metastatic disease [28], and very recently it was reported that KCNQ1 was a major CRC prognostic predictor classifier as CRC patients with stage II and stage III CRC who maintained expression of KCNQ1 showed a much stronger disease-free survival (DFS) [29].
Employing CRISPR/cas9 gene targeting an 11-bp insertion in the hamster KCNQ1 gene was introduced into the hamster genome, which resulted in a constitutive null allele [30]. KCNQ1 homozygous KO hamsters demonstrated similar neurological defects and other phenotypes that are similar to Kcnq1 KO mice [28], including inner ear defects, head bobbing and smaller stature [30]. Adult KCNQ1 homozygous KO hamsters developed severe physical stress as early as 70 days of age, including overt cancers at necropsy [30]. Overall, >85% of the KCNQ1 homozygous KO hamsters developed cancers, with the mean age when they became moribund at 150 days. None of the hamster littermate siblings that were either wild-type or heterozygous for KCNQ1 mutations developed cancers. The four most common cancers observed following pathological analysis were T-cell lymphomas (the most common cancer), plasma cell tumors, hemangiosarcomas, and myeloproliferative disease consistent with myeloid leukemia [30]. Figure 1 depicts gross and microscopic images of an intestinal T-cell lymphoma.
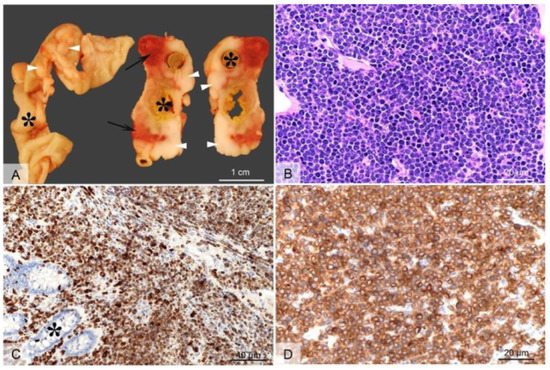
Figure 1. Gross and microscopic images of an intestinal T cell lymphoma in a KCNQ1 homozygous mutant hamster. Grossly (A), extending from the intestinal wall through the serosa and mesentery, there are multifocal to coalescing white-beige masses (arrow heads), with focal areas of necrosis and hemorrhage (arrows). The tumors circumferentially surround the intestines (indicated by asterisks) which are occasionally moderately dilated and segmentally ulcerated (transmural). Histologically, the tumor consists of densely packed sheets of round to oval cells with scant basophilic cytoplasm (B) that infiltrate the intestinal mucosa ((C), asterisk) and subjacent layers through to intestinal serosa. The cells show a uniform, intense immunolabeling of plasma membrane for CD3 (C,D) (anti-CD3 antibody, DAKO/Agilent, Santa Clara, CA, USA; Cat# A0452). H&E, 40× objective Panel (B); CD3 immunolabeling with 20× objective Panel (C), and with 40× objective Panel (D) (from [30], with permission).
The cancer phenotype in KCNQ1 mutant hamsters was unique in several aspects. First, the KCNQ1 KO hamsters represented the initial creation of a genetically engineered hamster cancer model. Second, while Kcnq1 mutant mice demonstrated an enhanced intestinal cancer phenotype when introgressed into the ApcMin model of intestinal cancer [28], Kcnq1 mutations alone in mice did not generate cancers in any tissue, unlike hamsters, who developed a range of cancers, sometimes multiple synchronous cancers, in different tissues. Similarly, humans who are mutant for KCNQ1 do not develop cancer, aside from the development of gastric hyperplasia which can represent a premalignant stage of gastric cancer. Thus, for reasons that remain unknown, the complete loss of KCNQ1 function uniquely leads to widespread blood cell cancer development in hamsters. How these cancers arise in KCNQ1 KO hamsters is an area of active investigation. Studies in humans and mice have shown that KCNQ1 is expressed in several types of hematopoietic cells, including thymic T cells, bone marrow, and other white blood cells, although KCNQ1 expression patterns in these cell types in hamsters have not been adequately characterized. Work by the group and others has indicated that KCNQ1 may be involved in the regulation of tissue stem cell transformation. This has been proposed in GI cancers associated with bidirectional Wnt/β-catenin signaling. Work by the group has shown that Kcnq1 mRNA [28] and protein expression (P. Scott et al., unpublished) was localized to the intestinal stem cell compartment. Further, Kcnq1 expression has been reported in a murine bone marrow-derived stem cell progenitor population. Ongoing studies are exploring whether Wnt/β-catenin signaling is dysregulated in KCNQ1-deficient bone marrow-derived hamster progenitor cells, of particular importance with the emerging evidence of a significant role for Wnt signaling in hematopoietic cancers.
3.2. TP53 Knockout Hamster Model
TP53 is considered the canonical tumor suppressor gene, commonly mutated in most major human cancers, and for this reason, it has been referred to as the “guardian of the genome” [31]. TP53 is a sequence-specific transcription factor that is involved in a range of cellular functions that are critical for its role in suppressing cancer, including cell cycle regulation, sensing of DNA damage, sensing and reaction to oxidative stress, apoptosis, senescence, and cellular metabolism [32]. Employing CRISPR/cas9 genetic engineering researchers created a 1-bp insertion in the hamster TP53 gene at amino acid position 311 that resulted in a truncation mutation that disrupts the DNA binding domain of TP53 [33]. Researchers found that both TP53 homozygous and TP53 heterozygous KO hamsters developed a wide range of cancers starting at 53 days of age, with individual hamsters often manifesting multiple synchronous cancers in different tissues. In contrast, a control group of TP53 wild-type siblings did not develop cancer, even when aged beyond one year. On average, TP53 homozygous mutants survived 139 days, and TP53 heterozygous mutants survived 286 days. Researchers analyzed by histopathology 52 TP53 homozygous mutant hamsters and 30 TP53 heterozygous mutant hamsters [33]. The homozygous mutant hamsters developed a wide range of cancers, with lymphomas (29%), hemangiosarcomas (27%), and myeloid leukemias (21%) representing the most common cancers. To a lesser extent, they developed several other sarcomas and adenocarcinomas in the adrenal glands, pancreas, and kidney. In TP53 heterozygous hamsters, lymphomas (67%) were the predominant type of cancer, followed by hemangiosarcomas (17%), myeloid leukemias (6%), and several other sarcomas and carcinomas that were found in only one hamster. Notably, in a group of 17 cancers from TP53 heterozygous animals all showed loss of heterozygosity (LOH) at the TP53 locus [33]. See Figure 2 depicts myeloid leukemia involvement in the liver of TP53 mutant hamsters.
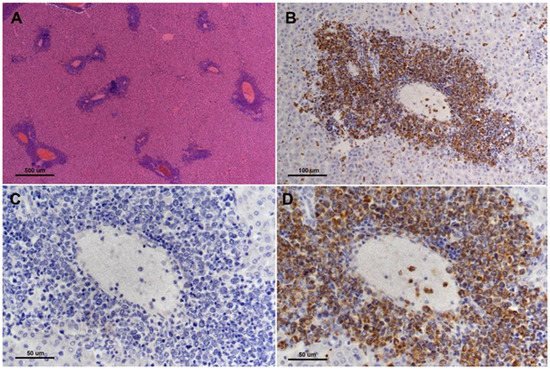
Figure 2. Myeloid leukemia involvement of liver of TP53 homozygous mutant hamster. There are multifocal infiltrates of neoplastic cells predominantly involving the portal triads ((A), H&E, 4× objective lens). Infiltrating cells surrounding hepatic portal veins are strongly myeloperoxidase positive ((B,D), 20× and 40× objective lens, respectively). (C) shows corresponding negative control (MPO-specific primary antibody is substituted with normal rabbit immunoglobulin on a serial step section, 40× objective lens) for image in (D) (from [33], with permission).
The cancer phenotypes in TP53 mutant hamsters were notable in several areas. First of all, in contrast with Trp53 homozygous KO mice, hamsters developed carcinomas in several epithelial tissues that were not observed in Trp53 KO mice. Human Li Fraumeni patients who carry germline TP53 mutations typically develop carcinomas while TP53 null mice primarily develop sarcomas. Trp53 mice carrying gain of function point mutations do develop a broader range of cancers that is closer to human cancers with TP53 mutations and TP53-deficient hamsters with one major exception in that they do not develop myeloid leukemias, which are common in TP53 mutants (especially homozygous mutant) hamsters and in human myeloid leukemia patients. TP53-deficient acute myelogenous leukemia (AML) is a particularly aggressive sub-type of AML with a median survival of 5–9 months and one-year survival of 0–10% [34]. Further, TP53 mutations are common in therapy-related AML (t-AML), a very aggressive disease because of the high likelihood of treatment failure. TP53 mutations are also found in 20–27% of AML with myelodysplasia-related changes (AML-MRC) and up to 40% of therapy-related AML myelodysplastic syndrome (AML/MDS) [34]. TP53 mutations are also associated with high-risk MDS and rapid transformation to secondary AML (sAML), an extremely lethal disease with less than 6 months median overall survival [34]. There are currently few treatment options for TP53-deficient AML and one of the major barriers to the development of new therapies has been the lack of a reliable rodent model. Trp53-deficient mice do not develop AML, but now there is a hamster model that can be used to study the genetic steps that cause this disease, including high-risk MDS that can rapidly transform to secondary AML. Hamsters can now also be used to test new drug modalities to treat TP53-deficient AML.
3.3. IL2RG KO Hamster Model
IL2RG encodes the commonly shared gamma chain subunit in the interleukin-2 receptors (IL2Rs) which meditate the functions of multiple type I interleukin cytokines, including interleukin-2 (IL-2), IL-4, IL-7, IL-9, IL-15 and IL-21 [35][36]. IL2RG resides on the X-chromosome and loss of function mutations in this gene cause X-linked severe combined immunodeficiency (XSCID) which is characterized by a failure in T development, lack of class switching recombination of immunoglobin genes in B lymphocytes, and the absence of natural killer (NK) cells. These severe immunodeficiency features highlight the importance of IL2RG in the development of adaptative immunity and innate immunity.
To develop a hamster model for XSCID, as well as a host for human tumor engraftment (discussed below), researchers recently created several IL2RG KO hamster lines. To genetically inactivate IL2RG in the hamster, researchers employed the CRISPR/cas9 system with single guide RNAs (sgRNAs) designed to target its exon 1. Among the produced hamster lines carrying different insertions and deletions (indels) in exon 1, lines with frameshift mutations causing multiple premature stop codons in IL2RG were chosen for the establishment of IL2RG KO hamster colonies and used to produce experimental animals [37][38]. Lymphocyte-specific gene expression analyses in the spleen showed that IL2RG KO hamsters present severe defects in lymphocyte development which are characterized by a greatly reduced number of CD4+ T cells and barely detectable CD8+ T cells, B cells, and NK cells, mimicking many aspects of human XSCID immunodeficiency. The use of these IL2RG KO hamsters as an XSCID model to study the infectious diseases that are of great health concern in immunocompromised patients was reported [36]. Here, researchers summarize the recent studies in using the IL2RG KO hamsters as a host for human tumor tissue engraftment to produce patient-derived xenograft (PDX) models as laboratory avatars for oncology research.
The use of immunodeficient animals, either with IL2RG-deficiency alone or in combination with other immunodeficiencies such as RAG2-deficiency, as hosts to produce PDX models relies on their incapability of mounting immune rejections to human tissues, therefore allowing them to be xenografted in the animal hosts. Compared to in vitro cultured tumor-derived cell lines where in vitro adaptation takes place which may change the physiology of tumors, the PDX models better recapitulate the tumor biology in vivo [38]. Il2rg KO mice have been widely used as hosts for producing PDX models [39]. However, there are several significant limitations in using mice as hosts for human tissue xenografting, chief among which are the incompatibility of mouse growth factors and cytokines with human tissues and cells, leading to low xenografting rates, limited or no self-renewal of malignant stem cells, and low cancer cell metastasis rates. The lack of functional communications between human cells/tissues and the mouse physiological milieu is also one of the underlying causes of the fundamental problems in using immunodeficient mice as hosts for human hematopoietic stem cell transplantation to produce so-called humanized mice carrying a humanized hematolymphoid system. Due to the lack of functional support from mouse cytokines for human lymphoid and myeloid cells, mice are incapable of fully supporting the development of a human immune system, as manifested by impaired lymph node development, poorly developed germinal centers, defective humoral immune responses (characterized by low levels of immunoglobulin production and inefficient immunoglobulin class switching recombination) and human NK cell development [40][41][42][43][44]. To address this species incompatibility issue, next-generation humanized mice transgenically expressing human cytokine genes, such as IL-3 and granulocyte macrophage-colony stimulating factor (GM-CSF) that are critical for myeloid cell development [45], and other human genes such as in the MISTRG mice [46], have been developed. The transgenic expression of these human genes has significantly improved the engraftment of human tumor tissues for mouse PDX model production [47] and the development of human lymphoid and myeloid cells. Nevertheless, these humanized mice are still not optimal in several aspects including being defective in mounting antigen-specific adaptive immune responses, low levels of reconstitution of gut-associated lymphoid tissues from human cells, and poor lymphoid architecture and organ development [42].
Therefore, a small rodent with better physiological compatibility with human cells/tissues is highly desired as a host for PDX and immune system humanization. In this regard, the golden Syrian hamster is a promising candidate, as in contrast to mice several human cytokines such as GM-CSF [48] and IL-12 [49][50] are cross-reactive with hamster cells. As mentioned above, human GM-CSF is among the key cytokines transgenically expressed in the MISTRG mice, which greatly improved human myeloid cell reconstitution [45]. To test whether the hamster is indeed a suitable host for human PDX, researchers carried out human pancreatic cancer cell transplantation experiments in the IL2RG KO hamsters and produced the first hamster PDX model. In these experiments, researchers used an IL2RG KO line (named ZZU001) that researchers generated in which a 10 bp frameshift deletion in exon 1 fully inactivated the expression of the IL2RG gene [37].
Researchers first performed subcutaneous xenotransplantation of a human pancreatic cancer cell line, MIA-PaCa-2 [51], into ZZU001 hamsters, and for comparisons, also into immunodeficient B-NDG (NOD-Prkdcscid IL2rgtm1/Bcgen) mice which are defective in both the IL2rg gene and the Prkdc gene. While xenografted human pancreatic cancer cells developed in both the hamster and B-NDG mouse hosts, no distant metastasis was identified in mice but was found in the lung, kidneys, and adrenal glands in ZZU001 hamsters. Figure 3 (From [37]) shows a H&E staining of a lung metastasis at 22 d, 29 d, and 36 d after subcutaneous injection of MIA-PaCa-2 cells into ZZU001 hamsters and B-NDG mice. The same observations, i.e., metastasis was only observed in the hamster hosts but not in the mouse hosts, were also made from each of the other four independent human pancreatic cancer cell lines tested.
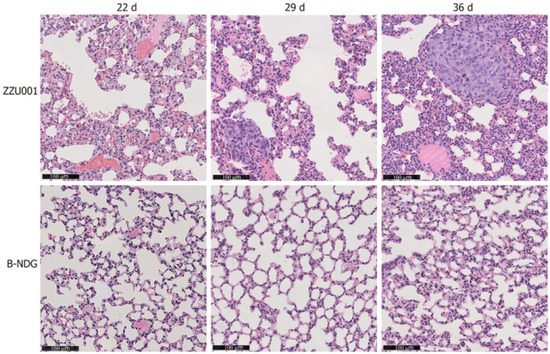
Figure 3. (From [37], with permission). H&E staining of a lung metastasis at 22 d, 29 d, 36 d after subcutaneous injection of MIA-PaCa-2 cells in ZZU001 hamsters and B-NDG mice. Scale bars = 100 µm.
Because orthotopic transplantation models mimic the biology of primary tumors more closely than subcutaneous transplantation models, to establish the feasibility of the hamster as an orthotopic model for human pancreatic cancer cell transplantation, orthotopic transplantation experiments with MIA-PaCa-2 cells were also performed in both ZZU001 hamsters and B-NDG mice. MIA-PaCa-2 cells were directly injected into the pancreas of hamsters and mice and tumor formation rates, as well as tumor metastasis, were compared between these two host species. Orthotopic transplantation of Mia-PaCa2 cells in hamsters led to a 100% (5/5) tumor formation rate and metastasis at multiple sites, including liver (100%), lung (100%), retroperitoneum (100%), mesentery (100%), kidney (100%), diaphragm (40%), adrenal gland (40%), and stomach (20%). In contrast, much lower metastatic rates to other organs were observed, with no lung metastasis observed at all, in the B-NDG hosts. In addition, metastatic tumors developed in the hamster orthotopic models presented similar clinical and pathological features to what was observed in human patients, which includes local infiltration of cancer cells, ascites, jaundice, ileus, and cachexia.
In summary, researchers demonstrated that in comparison to the highly immunodeficient B-NDG mice, ZZU001 hamsters with the loss of IL2RG function alone (therefore much less immunodeficient than B-NDG mice) support better human pancreatic cancer cell engraftment and multiple organ metastasis and serve as an improved host for human pancreatic cancer engraftment. Researchers posit that the improved engraftments and higher metastasis rates in the ZZU001 hamsters than in mouse hosts by human cancer cells are attributed to better communications between the human cancer cells with the hamster physiological milieu.
References
- Li, Z.; Xiong, C.; Mo, S.; Tian, H.; Yu, M.; Mao, T.; Chen, Q.; Luo, H.; Li, Q.; Lu, J.; et al. Comprehensive transcriptome analyses of the fructose-fed Syrian Golden hamster liver provides novel insights into lipid metabolism. PLoS ONE 2016, 11, e0162402.
- Ebihara, H.; Zivcec, M.; Gardner, D.; Falzarano, D.; LaCasse, R.; Rosenke, R.; Long, D.; Haddock, E.; Fischer, E.; Kawaoka, Y.; et al. A Syrian golden hamster model recapitulating ebola hemorrhagic fever. J. Infect. Dis. 2013, 207, 306–318.
- Guillaume, V.; Wong, K.T.; Looi, R.Y.; Georges-Courbot, M.C.; Barrot, L.; Buckland, R.; Wild, T.F.; Horvat, B. Acute Hendra virus infection: Analysis of the pathogenesis and passive antibody protection in the hamster model. Virology 2009, 387, 459–465.
- Jove, M.; Ayala, V.; Ramirez-Nunez, O.; Serrano, J.C.; Cassanye, A.; Arola, L.; Caimari, A.; Del Bas, J.M.; Crescenti, A.; Pamplona, R.; et al. Lipidomic and metabolomic analyses reveal potential plasma biomarkers of early atheromatous plaque formation in hamsters. Cardiovasc. Res. 2013, 97, 642–652.
- Paciello, O.; Wojcik, S.; Gradoni, L.; Oliva, G.; Trapani, F.; Iovane, V.; Politano, L.; Papparella, S. Syrian hamster infected with Leishmania infantum: A new experimental model for inflammatory myopathies. Muscle Nerve 2010, 41, 355–361.
- Best, E.L.; Freeman, J.; Wilcox, M.H. Models for the study of Clostridium difficile infection. Gut Microbes 2012, 3, 145–167.
- Wahl-Jensen, V.; Bollinger, L.; Safronetz, D.; de Kok-Mercado, F.; Scott, D.P.; Ebihara, H. Use of the Syrian hamster as a new model of ebola virus disease and other viral hemorrhagic fevers. Viruses 2012, 4, 3754–3784.
- Safronetz, D.; Ebihara, H.; Feldmann, H.; Hooper, J.W. The Syrian hamster model of hantavirus pulmonary syndrome. Antivir. Res. 2012, 95, 282–292.
- Weisburger, J.H. Colon carcinogens: Their metabolism and mode of action. Cancer 1971, 28, 60–70.
- Della Porta, G. Induction of intestinal, mammary, and ovarian tumors in hamsters with oral administration of 20-Methylcholanthrene. Cancer Res. 1961, 21, 575–578.
- Vairaktaris, E.; Spyridonidou, S.; Papakosta, V.; Vylliotis, A.; Lazaris, A.; Perrea, D.; Yapijakis, C.; Patsouris, E. The hamster model of sequential oral oncogenesis. Oral Oncol. 2008, 44, 315–324.
- Takahashi, M.; Hori, M.; Mutoh, M.; Wakabayashi, K.; Nakagama, H. Experimental animal models of pancreatic carcinogenesis for prevention studies and their relevance to human disease. Cancers 2011, 3, 582–602.
- Wold, W.S.; Toth, K. Chapter three—Syrian hamster as an animal model to study oncolytic adenoviruses and to evaluate the efficacy of antiviral compounds. Adv. Cancer Res. 2012, 115, 69–92.
- Ying, B.; Toth, K.; Spencer, J.F.; Aurora, R.; Wold, W.S.M. Transcriptome sequencing and development of an expression microarray platform for liver infection in adenovirus type 5-infected Syrian Golden hamsters. Virology 2015, 485, 305–312.
- Jin, J.; Guo, L.; VonTungeln, L.; Vanlandingham, M.; Cerniglia, C.E.; Chen, H. Smokeless tobacco impacts oral microbiota in a Syrian Golden hamster cheek pouch carcinogenesis model. Anaerobe 2018, 52, 29–42.
- Terasaki, M.; Nishizaka, Y.; Murase, W.; Kubota, A.; Kojima, H.; Kojoma, M.; Tanaka, T.; Maeda, H.; Miyashita, K.; Mutoh, M.; et al. Effect of Fucoxanthinol on Pancreatic Ductal Adenocarcinoma Cells from an N-Nitrosobis(2-oxopropyl)amine-initiated Syrian Golden Hamster Pancreatic Carcinogenesis Model. Cancer Genom. Proteom. 2021, 18 (Suppl. 3), 407–423.
- Wang, W.C.; Huang, M.Y.; Chen, Y.K.; Lan, W.C.; Shieh, T.M.; Shih, Y.H. Salivary Exosome Proteomics and Bioinformatics Analysis in 7,12-Dimethylbenzanthracene-Induced Oral Cancer with Radiation Therapy-A Syrian Golden Hamster Model. Diagnostics 2021, 12, 65.
- Kreimann, E.L.; Itoiz, M.E.; Dagrosa, A.; Garavaglia, R.; Farías, S.; Batistoni, D.; Schwint, A.E. The hamster cheek pouch as a model of oral cancer for boron neutron capture therapy studies: Selective delivery of boron by boronophenylalanine. Cancer Res. 2001, 61, 8775–8781.
- Santa Cruz, I.S.; Garabalino, M.A.; Trivillin, V.A.; Itoiz, M.E.; Pozzi, E.C.C.; Thorp, S.; Curotto, P.; Guidobono, J.S.; Heber, E.M.; Nigg, D.W.; et al. Optimization of the classical oral cancerization protocol in hamster to study oral cancer therapy. Oral Dis. 2020, 26, 1175–1184.
- Yapijakis, C.; Kalogera, S.; Papakosta, V.; Vassiliou, S. The Hamster Model of Sequential Oral Carcinogenesis: An Update. In Vivo 2019, 33, 1751–1755.
- Tchitchek, N.; Safronetz, D.; Rasmussen, A.L.; Martens, C.; Virtaneva, K.; Porcella, S.F.; Feldmann, H.; Ebihara, H.; Katze, M.G. Sequencing, annotation and analysis of the Syrian hamster (Mesocricetus auratus) transcriptome. PLoS ONE 2014, 9, e112617.
- Fan, Z.; Li, W.; Lee, S.R.; Meng, Q.; Shi, B.; Bunch, T.D.; White, K.L.; Kong, I.K.; Wang, Z. Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS ONE 2014, 9, e109755.
- Li, R.; Miao, J.; Fan, Z.; Song, S.; Kong, I.K.; Wang, Y.; Wang, Z. Production of Genetically Engineered Golden Syrian Hamsters by Pronuclear Injection of the CRISPR/Cas9 Complex. J. Vis. Exp. 2018, 131, e56263.
- Golden, J.W.; Li, R.; Cline, C.R.; Zeng, X.; Mucker, E.M.; Fuentes-Lao, A.J.; Spik, K.W.; Williams, J.A.; Twenhafel, N.; Davis, N.; et al. Hamsters Expressing Human Angiotensin-Converting Enzyme 2 Develop Severe Disease following Exposure to SARS-CoV-2. mBio 2022, 13, e0290621.
- Halfmann, P.J.; Kuroda, M.; Maemura, T.S.; Armbrust, T.; Wright, R.; Balaram, A.; Florek, K.R.; Bateman, A.C.; Kawaoka, Y. Efficacy of vaccination and previous infection against the Omicron BA.1 variant in Syrian hamsters. Cell Rep. 2022, 39, 110688.
- Uraki, R.; Kiso, M.; Iida, S.; Imai, M.; Takashita, E.; Kuroda, M.; Halfmann, P.J.; Loeber, S.; Maemura, T.; Yamayoshi, S.; et al. Characterization and antiviral susceptibility of SARS-CoV-2 Omicron BA.2. Nature 2022, 607, 119–127.
- den Uil, S.H.; Coupé, V.M.; Linnekamp, J.F.; van den Broek, E.; Goos, J.A.; Delis-van, D.P.M.; Belt, E.J.; van Grieken, N.C.; Scott, P.M.; Vermeulen, L.; et al. Loss of KCNQ1 expression in stage II and stage III colon cancer is a strong prognostic factor for disease recurrence. Br. J. Cancer 2016, 115, 1565–1574.
- Than, B.L.; Goos, J.A.; Sarver, A.L.; O’Sullivan, M.G.; Rod, A.; Starr, T.K.; Fijneman, R.J.; Meijer, G.A.; Zhao, L.; Zhang, Y.; et al. The role of KCNQ1 in mouse and human gastrointestinal cancers. Oncogene 2014, 33, 3861–3868.
- Uil, S.H.; Coupé, V.M.H.; Bril, H.; Meijer, G.A.; Fijneman, R.J.A.; Stockmann, H.B.A.C. KCNQ1 and lymphovascular invasion are key features in a prognostic classifier for stage II and III colon cancer. BMC Cancer 2022, 22, 372.
- Li, R.; Miao, J.; Tabaran, A.F.; O’Sullivan, M.G.; Anderson, K.J.; Scott, P.M.; Wang, Z.; Cormier, R.T. A novel cancer syndrome caused by KCNQ1-deficiency in the golden Syrian hamster. J. Carcinog. 2018, 17, 6.
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational landscape and significance across 12 major cancer types. Nature 2013, 502, 333–339.
- Aubrey, B.J.; Strasser, A. Tumor-Suppressor Functions of the TP53 Pathway. Cold Spring Harb. Perspect Med. 2016, 6, a026062.
- Miao, J.; Li, R.; Wettere, A.J.V.; Guo, H.; Tabaran, A.F.; Carlson, T.; Scott, P.M.; Chen, K.; Cormier, R.T. Cancer spectrum in TP53-deficient golden Syrian hamsters: A new model for Li-Fraumeni syndrome. J. Carcinog. 2021, 20, 18.
- Molica, M.; Mazzone, C.; Niscola, P.; de Fabritiis, P. TP53 Mutations in Acute Myeloid Leukemia: Still a Daunting Challenge? Front. Oncol. 2021, 10, 610820.
- Lin, J.X.; Leonard, W.J. The Common Cytokine Receptor γ Chain Family of Cytokines. Cold Spring Harb. Perspect Biol. 2018, 10, a028449.
- Li, R.; Ying, B.; Liu, Y.; Spencer, J.F.; Miao, J.; Tollefson, A.E.; Brien, J.D.; Wang, Y.; Wold, W.S.M.; Wang, Z.; et al. Generation and characterization of an IL2rg knockout Syrian hamster model for XSCID and HAdV-C6 infection in immunocompromised patients. Dis. Model Mech. 2020, 13, dmm044602.
- Miao, J.X.; Wang, J.Y.; Li, H.Z.; Guo, H.R.; Dunmall, L.S.C.; Zhang, Z.X.; Cheng, Z.G.; Gao, D.L.; Dong, J.Z.; Wang, Z.D.; et al. Promising xenograft animal model recapitulating the features of human pancreatic cancer. World J. Gastroenterol. 2020, 26, 4802–4816.
- Lan, T.; Xue, X.; Dunmall, L.C.; Miao, J.; Wang, Y. Patient-derived xenograft: A developing tool for screening biomarkers and potential therapeutic targets for human esophageal cancers. Aging 2021, 13, 12273–12293.
- Dobrolecki, L.E.; Airhart, S.D.; Alferez, D.G.; Aparicio, S.; Behbod, F.; Bentires-Alj, M.; Brisken, C.; Bult, C.J. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev. 2016, 35, 547–573.
- Walsh, N.C.; Kenney, L.L.; Jangalwe, S.; Aryee, K.E.; Greiner, D.L.; Brehm, M.A.; Shultz, L.D. Humanized Mouse Models of Clinical Disease. Annu. Rev. Pathol. 2017, 12, 187–215.
- Akkina, R.; Allam, A.; Balazs, A.B.; Blankson, J.N.; Burnett, J.C.; Casares, S.; Garcia, J.V.; Hasenkrug, K.J.; Kashanchi, F.; Kitchen, S.G.; et al. Improvements and Limitations of Humanized Mouse Models for HIV Research: NIH/NIAID “Meet the Experts” 2015 Workshop Summary. AIDS Res. Hum. Retrovir. 2016, 32, 109–119.
- Allen, T.M.; Brehm, M.A.; Bridges, S.; Ferguson, S.; Kumar, P.; Mirochnitchenko, O.; Palucka, K.; Pelanda, R.; Sanders-Beer, B.; Shultz, L.D.; et al. Humanized immune system mouse models: Progress, challenges and opportunities. Nat. Immunol. 2019, 20, 770–774.
- Shultz, L.D.; Lyons, B.L.; Burzenski, L.M.; Gott, B.; Chen, X.; Chaleff, S.; Kotb, M.; Gillies, S.D.; King, M.; Mangada, J.; et al. Human lymphoid and myeloid cell development in NOD/LtSz-scid IL2R gamma null mice engrafted with mobilized human hemopoietic stem cells. J. Immunol. 2005, 174, 6477–6489.
- Theocharides, A.P.; Rongvaux, A.; Fritsch, K.; Flavell, R.A.; Manz, M.G. Humanized hemato-lymphoid system mice. Haematologica 2016, 101, 5–19.
- Willinger, T.; Rongvaux, A.; Takizawa, H.; Yancopoulos, G.D.; Valenzuela, D.M.; Murphy, A.J.; Auerbach, W.; Eynon, E.E.; Stevens, S.; Manz, M.G.; et al. Human IL-3/GM-CSF knock-in mice support human alveolar macrophage development and human immune responses in the lung. Proc. Natl. Acad. Sci. USA 2011, 108, 2390–2395.
- Rongvaux, A.; Willinger, T.; Martinek, J.; Strowig, T.; Gearty, S.V.; Teichmann, L.L.; Saito, Y.; Marches, F.; Halene, S.; Palucka, A.K.; et al. Development and function of human innate immune cells in a humanized mouse model. Nat. Biotechnol. 2014, 32, 364–372.
- Song, Y.; Rongvaux, A.; Taylor, A.; Jiang, T.; Tebaldi, T.; Balasubramanian, K.; Bagale, A.; Terzi, Y.K.; Gbyli, R.; Wang, X.; et al. A highly efficient and faithful MDS patient-derived xenotransplantation model for pre-clinical studies. Nat. Commun. 2019, 10, 366.
- Cho, S.A.; Park, J.H.; Seok, S.H.; Juhn, J.H.; Kim, S.J.; Ji, H.J.; Choo, Y.S.; Park, J.H. Effect of granulocyte macrophage-colony stimulating factor (GM-CSF) on 5-FU-induced ulcerative mucositis in hamster buccal pouches. Exp. Toxicol. Pathol. 2006, 57, 321–328.
- Wang, P.; Li, X.; Wang, J.; Gao, D.; Li, Y.; Li, H.; Chu, Y.; Zhang, Z.; Liu, H.; Jiang, G.; et al. Re-designing Interleukin-12 to enhance its safety and potential as an anti-tumor immunotherapeutic agent. Nat. Commun. 2017, 8, 1395.
- Bortolanza, S.; Bunuales, M.; Otano, I.; Gonzalez-Aseguinolaza, G.; Ortiz-de-Solorzano, C.; Perez, D.; Prieto, J.; Hernandez-Alcoceba, R. Treatment of pancreatic cancer with an oncolytic adenovirus expressing interleukin-12 in Syrian hamsters. Mol. Ther. 2009, 17, 614–622.
- Yunis, A.A.; Arimura, G.K.; Russin, D.J. Human pancreatic carcinoma (MIA PaCa-2) in continuous culture: Sensitivity to asparaginase. Int. J. Cancer 1977, 19, 128–135.
More
Information
Subjects:
Oncology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.0K
Revisions:
2 times
(View History)
Update Date:
19 Aug 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




