Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Saskya Carrera | -- | 2031 | 2022-07-29 14:35:35 | | | |
| 2 | Dean Liu | Meta information modification | 2031 | 2022-08-01 03:56:18 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Barba-Ostria, C.; Carrera-Pacheco, S.E.; Gonzalez-Pastor, R.; Heredia-Moya, J.; Mayorga-Ramos, A.; Rodríguez-Pólit, C.; Zúñiga-Miranda, J.; Arias-Almeida, B.; Guamán, L.P. Evaluation of Biological Activity of Natural Compounds. Encyclopedia. Available online: https://encyclopedia.pub/entry/25666 (accessed on 08 February 2026).
Barba-Ostria C, Carrera-Pacheco SE, Gonzalez-Pastor R, Heredia-Moya J, Mayorga-Ramos A, Rodríguez-Pólit C, et al. Evaluation of Biological Activity of Natural Compounds. Encyclopedia. Available at: https://encyclopedia.pub/entry/25666. Accessed February 08, 2026.
Barba-Ostria, Carlos, Saskya E. Carrera-Pacheco, Rebeca Gonzalez-Pastor, Jorge Heredia-Moya, Arianna Mayorga-Ramos, Cristina Rodríguez-Pólit, Johana Zúñiga-Miranda, Benjamin Arias-Almeida, Linda P. Guamán. "Evaluation of Biological Activity of Natural Compounds" Encyclopedia, https://encyclopedia.pub/entry/25666 (accessed February 08, 2026).
Barba-Ostria, C., Carrera-Pacheco, S.E., Gonzalez-Pastor, R., Heredia-Moya, J., Mayorga-Ramos, A., Rodríguez-Pólit, C., Zúñiga-Miranda, J., Arias-Almeida, B., & Guamán, L.P. (2022, July 29). Evaluation of Biological Activity of Natural Compounds. In Encyclopedia. https://encyclopedia.pub/entry/25666
Barba-Ostria, Carlos, et al. "Evaluation of Biological Activity of Natural Compounds." Encyclopedia. Web. 29 July, 2022.
Copy Citation
Natural compounds have diverse structures and are present in different forms of life. Metabolites such as tannins, anthocyanins, and alkaloids, among others, serve as a defense mechanism in live organisms and are undoubtedly compounds of interest for the food, cosmetic, and pharmaceutical industries.
natural product
bioactive compounds
antimicrobial
antioxidant
1. Introduction
According to the World Health Organization (WHO), 80% of the global population uses medicinal-plant-based medicine to alleviate or cure diseases [1]. In addition, although estimates vary depending on what is considered a natural-product-derived drug, it is safe to say that up to 50% of currently marketed drugs owe their origins to natural products [2]. New molecules from natural resources with potential bioactivity are reported every day; however, only a few of these molecules are evaluated for their suitability for use as drugs [3]. Identifying bioactive compounds (hits or leads) is the initial step for drug discovery. Therefore, it is necessary to select suitable bioassays to evaluate both the activity against the disease and the potency. For this purpose, target-based screening is mainly used to identify compounds that modulate the activity of a target involved in a disease. This screening involves different in vitro biological assays designed to measure primary activities, selectivity, cellular toxicity, and physiologically relevant activity. The initial phases of a target-based screening cascade typically employ a range of in vitro assays, especially high-throughput screening (HTS); however, the study is more expensive and time-consuming [4][5]. In the first instance, the assays can be selected considering that structurally similar compounds have similar biological activity; however, this cannot always be carried out, especially when working with natural compounds or natural extracts.`
Extracts from various plants are commonly used in traditional medicine, either alone or in combination, but in many cases only a few of them are evaluated for their biological activity. However, due to inadequate fractionation processes or the degradation of active compounds during separation, it is not always easy to identify the molecules responsible for the activity of these extracts. Since obtaining an isolated compound very often requires infrastructure and specialized personnel and is expensive, it is challenging to offer pharmacological alternatives of this nature to people with low incomes. In addition, since medicinal herbs can be grown locally at a reasonable cost, natural extracts remain an option for treating some diseases in populations with limited resources or living in remote areas [6].
An important advantage of using crude extracts vs. isolated molecules is the presence of molecules in the extract that can interact synergistically with the bioactive compound, potentiating its beneficial effect [7]. However, despite their potential as accessible treatment options and as sources of bioactive molecules for drug discovery, it must be highlighted that similar to other pharmacological alternatives, natural extracts can also present adverse effects that should be considered and evaluated. Having many assays on hand to discard or confirm activities is critical when looking for bioactive compounds because the aim while looking for therapeutic agents is to identify an acceptable technique that can screen the source material for bioactivity.
2. Cytotoxicity Activity in Cultured Mammalian Cells
In the early stages of drug development, extensive toxicity screening is essential [8][9]. Animal studies involve high costs and are often restricted by differential responses due to the physiological differences between species and limitations in test feasibility. Alternatively, in vitro cytotoxicity assays are advantageous in preclinical studies due to their eligibility, cost-effectiveness, and reproducibility. Natural compounds have become particularly relevant in identifying safer and more effective treatments [10].
To evaluate the cytotoxicity in mammalian cells, it is critical to select the proper cell line for each particular experiment, considering the relevant species, specific organs, and chosen route of administration. Multiple cell lines are available for in vitro testing, including immortalized cell lines, primary cultures, and stem cells. Each cell line has requirements that need to be defined before the experiment to ensure a proper understanding of the potential mechanisms of toxicity [11][12].
A wide range of in vitro assays are currently available for cytotoxicity testing [13][14]. While direct cytotoxicity assays focus on detecting a loss of membrane integrity associated with cell death, cell viability assays are developed to measure the activity related to cellular maintenance and survival. Additionally, other assays allow the direct and indirect quantification of changes in the population at specific phases of the cell cycle and provide information on the mechanism of cell death [15]. Different approaches are employed in parallel to better understand the cytotoxicity mechanisms, such as the terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) assay and the comet assay for the analysis of DNA fragmentation [16]; the determination of the activation of apoptosis-related caspases [17][18]; or the detection of the relative level of telomerase activity (TRAP, telomerase repeat amplification protocol) [19][20]. Determining the cytotoxicity against tumor cells; detecting the rate and regulation of cell migration; and analyzing anchorage-independent proliferation, chemotaxis, and invasion are essential factors usually evaluated using colony-forming assays in soft agar and scratch assays using dual-chamber systems [21][22][23].
In short, each assay has its limitations. Although the 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) assay has been generally accepted as the gold standard in cytotoxicity testing, this method is not always the most pertinent due to interference with the test compounds, particularly with natural extracts [24][25]. Therefore, before selecting an appropriate methodology, different assays should be compared and more than one should be used when possible. The cost, reliability, timing, user-friendliness, and equipment requirements should also be considered [26].
Although two-dimensional mammalian monocultures stemming from specific cell types are widely used based on their reproducible and rapid growth, high productivity levels, ease of data interpretation, and value, the artificial nature of the culture environment presents limitations in drug safety and efficacy evaluation [27][28]. In this sense, even though improved versions of the classical reagents are being developed, the field’s current focus is shifting towards co-culture systems, human organoids, and other sophisticated three-dimensional culture models that collect more physiologically relevant data and represent methods that could better connect traditional cell culture and in vivo models [29][30][31] (Figure 1).
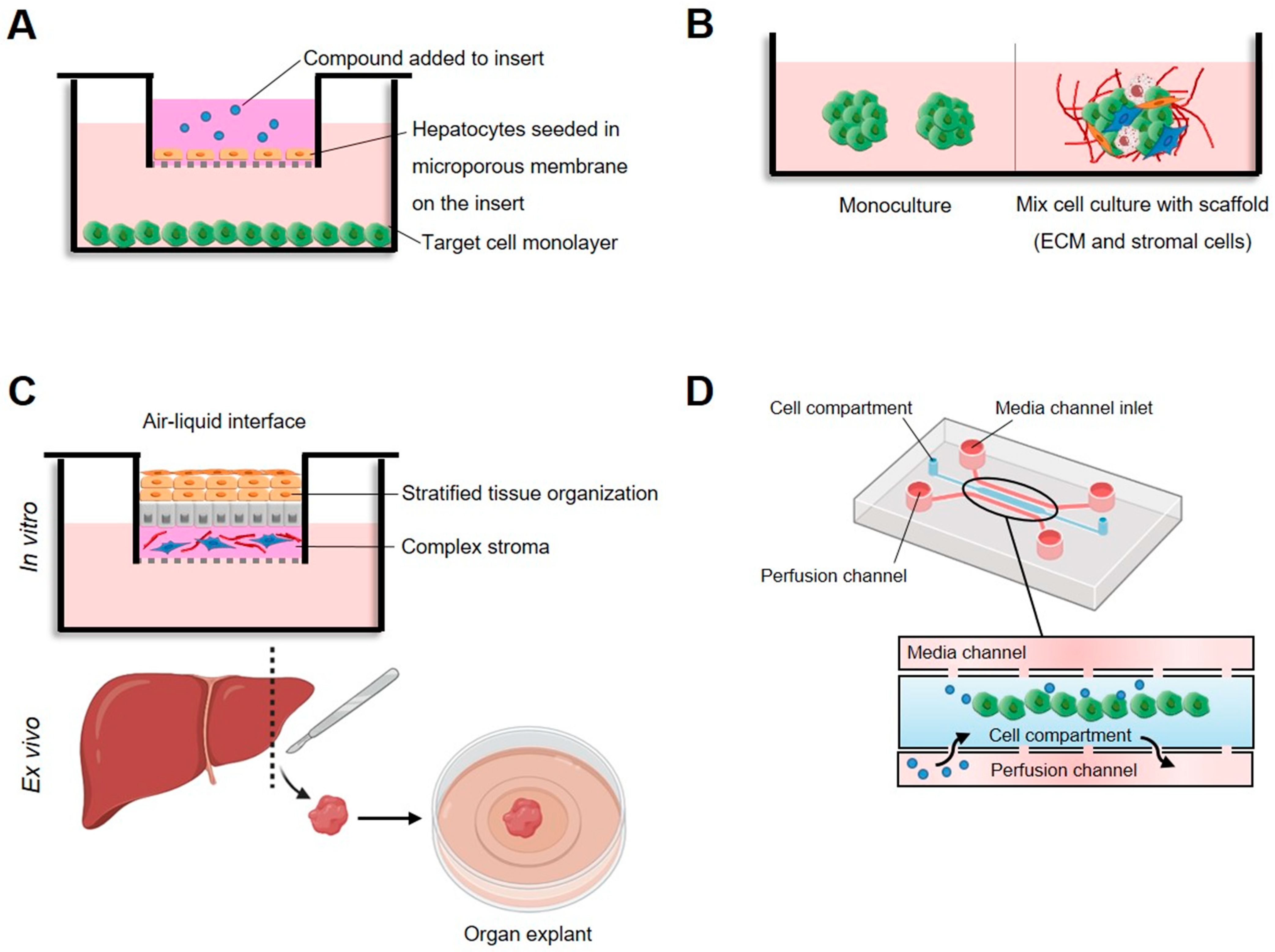
Figure 1. High-tech in vitro models to assess cytotoxicity in cultured mammalian cells. (A) Dual chamber, test compound, and metabolites diffuse through the microporous barrier toward target cells. (B) Three-dimensional cellular models based on multicellular spheroids or organoids consisting of target cells or the co-cultivation of several types of cells on extracellular matrix (ECM). (C) Organotypic cultures, whereby cells, organ slices, or whole organs are cultured on a tissue culture insert that is either submerged in medium or maintained at an air–liquid interface to ensure sufficient oxygen supply. (D) Microfluidic system based on a mixture of cells and matrix collected in the central channel and medium flowing from the lateral channels that keeps particles in homogenous suspension.
From the array of cell cultures available for in vitro testing that offer diverse degrees of intricacy and similarity to the in vivo setting, organotypic cultures are tissue slices that maintain cell interactions and the extracellular matrix composition of the original tissue and tissue function [32][33][34][35]. However, this system lacks intercommunication with the circulatory and immune systems and is inadequate for medium- to high-throughput analysis [11]. Three-dimensional spheroids and organoids self-organize into organ-specific structures that accurately replicate paracrine and direct intercellular interactions [36][37][38]. While spheroids are usually made from cell lines and offer lower complexity, organoids are derived from the stem cells of different origins and resemble the original tissue in the structure, histologically and genetically [39][40]. Tumor organoids are particularly relevant, since these systems provide suitable platforms to recapitulate the complex tumor microenvironment and its heterogeneity, allowing the study of chemical and metabolic gradients and mechanisms of resistance [29][41].
The evidence indicates that microfluidic devices are gaining traction in the area of cytotoxicity assays [42][43]. Compared to static conditions, microfluidic systems can reproduce the specific flow, temperature, pressure, and chemical gradients of the in vivo systems [44][45][46]. Thus, they can reconstruct the continuous renewal of nutrients, gasses and toxic wastes, migration, and microcirculation. These systems also support longer culture times and drug treatments that are more pharmacologically significant [47].
3. Antihyperglycemic Activity
Diabetes is a global health disease affecting 422 million people worldwide [48]. This disease is characterized by elevated blood glucose levels, which if untreated leads to severe multi-organ failure and 1.5 million deaths each year. Antihyperglycemic agents are a heterogeneous group of molecules obtained via chemical synthesis or isolation from natural sources that lower the glucose concentration in the blood or prevent its increase [49].
(i) assays based on the inhibition of isolated enzymes involved in the regulation of blood glucose levels and (ii) assays used to measure major cellular processes that directly alter glucose levels, mainly glucose uptake and insulin secretion [50]. The first group includes enzymes that catalyze the breakdown of poly- and oligosaccharides such as α-amylase and α-glucosidase, respectively. The inhibition of the mentioned enzymes and others with a similar role in carbohydrate digestion is considered antidiabetic. The reduction in the glucose concentration available to absorb in the intestine prevents a further increase in blood glucose [51]. The α-amylase and α-glucosidase inhibition assays are reactions of commercially available enzymes and substrates in the optimal conditions (buffer, pH, cofactors), allowing the detection of the reaction product(s) [52].
The most common substrate used to measure α-amylase is starch. The method is based on the reaction of starch with dinitrosalicylic acid (DNS), which reacts with reducing sugars, producing 3-amino-5-nitrosalicylic acid, which is measured spectrophotometrically at 540 nm. Most α-glucosidase assays rely on the spectrophotometric detection of p-nitrophenol liberated after the hydrolysis of p-nitrophenyl-α-d-glucopyranoside (pNPG), which can be measured at 400 nm. Dipeptidyl peptidase IV (DPP4) and tyrosine phosphatase 1B (TP1B) are involved in the indirect regulation of glucose levels by modulating insulin secretion (DPP4) [53] and signaling (TP1B) [54], respectively. DPP4 is a serine exopeptidase that cleaves different peptides, including GLP-1, a major regulator of insulin secretion in response to glucose [55]. Thus, inhibiting DPP-4 in vivo increases the availability of GLP-1 and insulin secretion, reducing blood glucose [56]. TP1B negatively regulates insulin and leptin signaling by dephosphorylating the insulin receptor (IR) and its downstream signaling components [57][58]. The inhibition of TP1B releases insulin signaling from TP1B-mediated dephosphorylation and allows insulin downstream signaling [52]. The second group includes assays designed to measure the potential inhibitory effects of different molecules on relevant cellular processes controlling blood glucose levels, such as glucose uptake and insulin secretion. The glucose uptake assay is based on the internalization of a labeled glucose analog that cannot be fully utilized because of its modification. It accumulates inside the cells, facilitating its detection. The output generated by the accumulation of labeled analogs is proportional to the glucose uptake and can be detected and quantified using standard equipment such as fluorescence or bioluminescence readers [59][60] or fluorescence-activated cell sorting (FACS) [61]. These assays can be performed in mammalian cell lines, given the importance of measuring the physiologically relevant effects. Some studies report using yeast cells as an alternative to mammalian cell lines [62]. For example, a recent study described a label-free method to measure glucose uptake in yeast cells using pHluorin, a genetically encoded pH-sensitive green fluorescent protein [63]. In general, insulin secretion assays are performed using ß-cells isolated from pancreatic or islet cell cultures. The cells are stimulated by glucose and incubated with the compound or plant extract to measure the insulin secretion modulation effect [64]. After being released from cells, insulin can be measured via radioimmunoassay [65][66] or ELISA. Recently, a luminescent alternative to detect insulin was described by Hager et al. and Kalwat et al. [67][68].
Despite continuous improvements in measuring glucose and glucose-associated processes, a relevant challenge is to fully understand the physiological and pathological roles of blood glucose levels and their impact on health conditions and disease. In vitro methods are critical for discovering natural compounds with antidiabetic activity. Although diabetes and other health issues associated with high blood glucose levels can be treated using available antidiabetics, given the vast chemical diversity of natural products with unknown but potentially beneficial effects, the evaluation of the antidiabetic activity of molecules obtained from natural sources is a very relevant research topic. Some parameters that are actively being improved to allow the efficient prospection of potential candidates for developing novel antidiabetic drugs from natural sources are: (i) increasing the sensitivity and robustness of the assays; (ii) reducing the laborious steps needed for the preparation of samples and cell extracts; and (iii) increasing the throughput of the assays.
References
- Malaquias, G.; Santos Cerqueira, G.; Pinheiro Ferreira, P.M.; Landim Pacheco, A.C.; de Castro e Souza, J.M.; do Socorro Meireles de Deus, M.; Peron, A.P. Utilização na medicina popular, potencial terapêutico e toxicidade em nível celular das plantas Rosmarinus officinalis L., Salvia officinalis L. e Mentha piperita L. (Família Lamiaceae). Rev. Intertox Toxicol. Risco Ambient. Soc. 2015, 7, 50–68.
- Kingston, D.G.I. Modern natural products drug discovery and its relevance to biodiversity conservation. J. Nat. Prod. 2011, 74, 496–511.
- Fabricant, D.S.; Farnsworth, N.R. The value of plants used in traditional medicine for drug discovery. Environ. Health Perspect. 2001, 109 (Suppl. S1), 69–75.
- Batool, M.; Ahmad, B.; Choi, S. A Structure-Based Drug Discovery Paradigm. Int. J. Mol. Sci. 2019, 20, 2783.
- Martin, Y.C.; Kofron, J.L.; Traphagen, L.M. Do structurally similar molecules have similar biological activity? J. Med. Chem. 2002, 45, 4350–4358.
- Rasoanaivo, P.; Wright, C.W.; Willcox, M.L.; Gilbert, B. Whole plant extracts versus single compounds for the treatment of malaria: Synergy and positive interactions. Malar. J. 2011, 10 (Suppl. S1), S4.
- Gilbert, B.; Alves, L. Synergy in plant medicines. Curr. Med. Chem. 2003, 10, 13–20.
- Vinken, M.; Blaauboer, B.J. In vitro testing of basal cytotoxicity: Establishment of an adverse outcome pathway from chemical insult to cell death. Toxicol. Vitr. 2017, 39, 104–110.
- Di Nunzio, M.; Valli, V.; Tomás-Cobos, L.; Tomás-Chisbert, T.; Murgui-Bosch, L.; Danesi, F.; Bordoni, A. Is cytotoxicity a determinant of the different in vitro and in vivo effects of bioactives? BMC Complement. Altern. Med. 2017, 17, 453.
- Ling, T.; Lang, W.H.; Maier, J.; Quintana Centurion, M.; Rivas, F. Cytostatic and Cytotoxic Natural Products against Cancer Cell Models. Molecules 2019, 24, 2012.
- Sachana, M.; Hargreaves, A.J. Chapter 9—Toxicological testing: In vivo and in vitro models. In Veterinary Toxicology, 3rd ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 145–161.
- Jablonská, E.; Kubásek, J.; Vojtěch, D.; Ruml, T.; Lipov, J. Test conditions can significantly affect the results of in vitro cytotoxicity testing of degradable metallic biomaterials. Sci. Rep. 2021, 11, 6628.
- Kamiloglu, S.; Sari, G.; Ozdal, T.; Capanoglu, E. Guidelines for cell viability assays. Food Front. 2020, 1, 332–349.
- Riss, T.; Niles, A.; Moravec, R.; Karassina, N.; Vidugiriene, J. Cytotoxicity assays: In vitro methods to measure dead cells. In Assay Guidance Manual; Sittampalam, G.S., Coussens, N.P., Brimacombe, K., Grossman, A., Arkin, M., Auld, D., Austin, C., Baell, J., Bejcek, B., Caaveiro, J.M.M., et al., Eds.; Eli Lilly & Company and the National Center for Advancing Translational Sciences: Bethesda, MD, USA, 2004.
- Gordon, J.L.; Brown, M.A.; Reynolds, M.M. Cell-Based Methods for Determination of Efficacy for Candidate Therapeutics in the Clinical Management of Cancer. Diseases 2018, 6, 85.
- King, T.C. Cell injury, cellular responses to injury, and cell death. In Elsevier’s Integrated Pathology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 1–20. ISBN 9780323043281.
- McStay, G.P.; Green, D.R. Measuring apoptosis: Caspase inhibitors and activity assays. Cold Spring Harb. Protoc. 2014, 2014, 799–806.
- Elmore, S. Apoptosis: A review of programmed cell death. Toxicol. Pathol. 2007, 35, 495–516.
- Heller-Uszynska, K.; Kilian, A. Microarray TRAP—A high-throughput assay to quantitate telomerase activity. Biochem. Biophys. Res. Commun. 2004, 323, 465–472.
- Menyhárt, O.; Harami-Papp, H.; Sukumar, S.; Schäfer, R.; Magnani, L.; de Barrios, O.; Győrffy, B. Guidelines for the selection of functional assays to evaluate the hallmarks of cancer. Biochim. Biophys. Acta 2016, 1866, 300–319.
- Borowicz, S.; Van Scoyk, M.; Avasarala, S.; Karuppusamy Rathinam, M.K.; Tauler, J.; Bikkavilli, R.K.; Winn, R.A. The soft agar colony formation assay. J. Vis. Exp. 2014, 92, e51998.
- Hulkower, K.I.; Herber, R.L. Cell migration and invasion assays as tools for drug discovery. Pharmaceutics 2011, 3, 107–124.
- Grada, A.; Otero-Vinas, M.; Prieto-Castrillo, F.; Obagi, Z.; Falanga, V. Research techniques made simple: Analysis of collective cell migration using the wound healing assay. J. Investig. Dermatol. 2017, 137, e11–e16.
- van Tonder, A.; Joubert, A.M.; Cromarty, A.D. Limitations of the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl-2H-tetrazolium bromide (MTT) assay when compared to three commonly used cell enumeration assays. BMC Res. Notes 2015, 8, 47.
- Akter, S.; Addepalli, R.; Netzel, M.E.; Tinggi, U.; Fletcher, M.T.; Sultanbawa, Y.; Osborne, S.A. Antioxidant-Rich Extracts of Terminalia ferdinandiana Interfere with Estimation of Cell Viability. Antioxidants 2019, 8, 191.
- Niles, A.L.; Moravec, R.A.; Riss, T.L. Update on in vitro cytotoxicity assays for drug development. Expert Opin. Drug Discov. 2008, 3, 655–669.
- Ballav, S.; Jaywant Deshmukh, A.; Siddiqui, S.; Aich, J.; Basu, S. Two-Dimensional and Three-Dimensional Cell Culture and Their Applications. In Cell Culture—Advanced Technology and Applications in Medical and Life Sciences; Biochemistry; IntechOpen: London, UK, 2021.
- Kapałczyńska, M.; Kolenda, T.; Przybyła, W.; Zajączkowska, M.; Teresiak, A.; Filas, V.; Ibbs, M.; Bliźniak, R.; Łuczewski, Ł.; Lamperska, K. 2D and 3D cell cultures—A comparison of different types of cancer cell cultures. Arch. Med. Sci. 2018, 14, 910–919.
- Jensen, C.; Teng, Y. Is it time to start transitioning from 2D to 3D cell culture? Front. Mol. Biosci. 2020, 7, 33.
- Hoarau-Véchot, J.; Rafii, A.; Touboul, C.; Pasquier, J. Halfway between 2D and Animal Models: Are 3D Cultures the Ideal Tool to Study Cancer-Microenvironment Interactions? Int. J. Mol. Sci. 2018, 19, 181.
- Berrouet, C.; Dorilas, N.; Rejniak, K.A.; Tuncer, N. Comparison of Drug Inhibitory Effects (IC50) in Monolayer and Spheroid Cultures. Bull. Math. Biol. 2020, 82, 68.
- Martin, S.Z.; Wagner, D.C.; Hörner, N.; Horst, D.; Lang, H.; Tagscherer, K.E.; Roth, W. Ex vivo tissue slice culture system to measure drug-response rates of hepatic metastatic colorectal cancer. BMC Cancer 2019, 19, 1030.
- Roelants, C.; Pillet, C.; Franquet, Q.; Sarrazin, C.; Peilleron, N.; Giacosa, S.; Guyon, L.; Fontanell, A.; Fiard, G.; Long, J.-A.; et al. Ex-Vivo Treatment of Tumor Tissue Slices as a Predictive Preclinical Method to Evaluate Targeted Therapies for Patients with Renal Carcinoma. Cancers 2020, 12, 232.
- Koerfer, J.; Kallendrusch, S.; Merz, F.; Wittekind, C.; Kubick, C.; Kassahun, W.T.; Schumacher, G.; Moebius, C.; Gaßler, N.; Schopow, N.; et al. Organotypic slice cultures of human gastric and esophagogastric junction cancer. Cancer Med. 2016, 5, 1444–1453.
- Temblador, A.; Topalis, D.; van den Oord, J.; Andrei, G.; Snoeck, R. Organotypic Epithelial Raft Cultures as a Three-Dimensional In Vitro Model of Merkel Cell Carcinoma. Cancers 2022, 14, 1091.
- Lin, R.-Z.; Lin, R.-Z.; Chang, H.-Y. Recent advances in three-dimensional multicellular spheroid culture for biomedical research. Biotechnol. J. 2008, 3, 1172–1184.
- Cui, X.; Hartanto, Y.; Zhang, H. Advances in multicellular spheroids formation. J. R. Soc. Interface 2017, 14, 20160877.
- Shankaran, A.; Prasad, K.; Chaudhari, S.; Brand, A.; Satyamoorthy, K. Advances in development and application of human organoids. 3 Biotech 2021, 11, 257.
- Caipa Garcia, A.L.; Arlt, V.M.; Phillips, D.H. Organoids for toxicology and genetic toxicology: Applications with drugs and prospects for environmental carcinogenesis. Mutagenesis 2022, 37, 143–154.
- Fey, S.J.; Wrzesinski, K. Determination of drug toxicity using 3D spheroids constructed from an immortal human hepatocyte cell line. Toxicol. Sci. 2012, 127, 403–411.
- Weeber, F.; Ooft, S.N.; Dijkstra, K.K.; Voest, E.E. Tumor Organoids as a Pre-clinical Cancer Model for Drug Discovery. Cell Chem. Biol. 2017, 24, 1092–1100.
- Kitaeva, K.V.; Rutland, C.S.; Rizvanov, A.A.; Solovyeva, V.V. Cell Culture Based in vitro Test Systems for Anticancer Drug Screening. Front. Bioeng. Biotechnol. 2020, 8, 322.
- Garcia-Hernando, M.; Benito-Lopez, F.; Basabe-Desmonts, L. Advances in microtechnology for improved cytotoxicity assessment. Front. Mater. 2020, 7, 582030.
- Mi, S.; Du, Z.; Xu, Y.; Wu, Z.; Qian, X.; Zhang, M.; Sun, W. Microfluidic co-culture system for cancer migratory analysis and anti-metastatic drugs screening. Sci. Rep. 2016, 6, 35544.
- Bhatia, S.N.; Ingber, D.E. Microfluidic organs-on-chips. Nat. Biotechnol. 2014, 32, 760–772.
- Bhise, N.S.; Ribas, J.; Manoharan, V.; Zhang, Y.S.; Polini, A.; Massa, S.; Dokmeci, M.R.; Khademhosseini, A. Organ-on-a-chip platforms for studying drug delivery systems. J. Control. Release 2014, 190, 82–93.
- Lubamba, B.; Jensen, T.; McClelland, R. Rapid Detection of Direct Compound Toxicity and Trailing Detection of Indirect Cell Metabolite Toxicity in a 96-Well Fluidic Culture Device for Cell-Based Screening Environments: Tactics in Six Sigma Quality Control Charts. Appl. Sci. 2022, 12, 2786.
- Roglic, G.; World Health Organization. Global Report on Diabetes; World Health Organization: Geneva, Switzerland, 2016; ISBN 9789241565257.
- Reyes, B.A.S.; Dufourt, E.C.; Ross, J.; Warner, M.J.; Tanquilut, N.C.; Leung, A.B. Selected phyto and marine bioactive compounds: Alternatives for the treatment of type 2 diabetes. In Studies in Natural Products Chemistry; Elsevier: Amsterdam, The Netherlands, 2017; Volume 55, pp. 111–143. ISBN 9780444640680.
- Thete, M.; Dilip, A. Recent advances and methods for in-vitro evaluation of antidiabetic activity: A review. Int. J. Eng. Appl. Sci. Technol. 2020, 4, 194–198.
- Gromova, L.V.; Fetissov, S.O.; Gruzdkov, A.A. Mechanisms of glucose absorption in the small intestine in health and metabolic diseases and their role in appetite regulation. Nutrients 2021, 13, 2474.
- Xue, B.; Kim, Y.-B.; Lee, A.; Toschi, E.; Bonner-Weir, S.; Kahn, C.R.; Neel, B.G.; Kahn, B.B. Protein-tyrosine phosphatase 1B deficiency reduces insulin resistance and the diabetic phenotype in mice with polygenic insulin resistance. J. Biol. Chem. 2007, 282, 23829–23840.
- Deacon, C.F. Physiology and Pharmacology of DPP-4 in Glucose Homeostasis and the Treatment of Type 2 Diabetes. Front. Endocrinol. 2019, 10, 80.
- Prabhakar, P.K.; Sivakumar, P.M. Protein Tyrosine Phosphatase 1B Inhibitors: A Novel Therapeutic Strategy for the Management of type 2 Diabetes Mellitus. Curr. Pharm. Des. 2019, 25, 2526–2539.
- Mulvihill, E.E.; Drucker, D.J. Pharmacology, physiology, and mechanisms of action of dipeptidyl peptidase-4 inhibitors. Endocr. Rev. 2014, 35, 992–1019.
- Omar, B.; Ahrén, B. Pleiotropic mechanisms for the glucose-lowering action of DPP-4 inhibitors. Diabetes 2014, 63, 2196–2202.
- Yip, S.-C.; Saha, S.; Chernoff, J. PTP1B: A double agent in metabolism and oncogenesis. Trends Biochem. Sci. 2010, 35, 442–449.
- Elchebly, M.; Payette, P.; Michaliszyn, E.; Cromlish, W.; Collins, S.; Loy, A.L.; Normandin, D.; Cheng, A.; Himms-Hagen, J.; Chan, C.C.; et al. Increased insulin sensitivity and obesity resistance in mice lacking the protein tyrosine phosphatase-1B gene. Science 1999, 283, 1544–1548.
- Csepregi, R.; Temesfői, V.; Sali, N.; Poór, M.; Needs, P.W.; Kroon, P.A.; Kőszegi, T. A One-Step Extraction and Luminescence Assay for Quantifying Glucose and ATP Levels in Cultured HepG2 Cells. Int. J. Mol. Sci. 2018, 19, 2670.
- Blodgett, A.B.; Kothinti, R.K.; Kamyshko, I.; Petering, D.H.; Kumar, S.; Tabatabai, N.M. A fluorescence method for measurement of glucose transport in kidney cells. Diabetes Technol. Ther. 2011, 13, 743–751.
- Dong, S.; Alahari, S. FACS-based Glucose Uptake Assay of Mouse Embryonic Fibroblasts and Breast Cancer Cells Using 2-NBDG Probe. Bio Protoc. 2018, 8, e2816.
- Cirillo, V.P. Mechanism of glucose transport across the yeast cell membrane. J. Bacteriol. 1962, 84, 485–491.
- Schmidl, S.; Iancu, C.V.; Reifenrath, M.; Choe, J.-Y.; Oreb, M. A label-free real-time method for measuring glucose uptake kinetics in yeast. FEMS Yeast Res. 2021, 21, foaa069.
- Lee, J.; Noh, S.; Lim, S.; Kim, B. Plant extracts for type 2 diabetes: From traditional medicine to modern drug discovery. Antioxidants 2021, 10, 81.
- Schmidt, S.; Jakab, M.; Jav, S.; Streif, D.; Pitschmann, A.; Zehl, M.; Purevsuren, S.; Glasl, S.; Ritter, M. Extracts from Leonurus sibiricus L. increase insulin secretion and proliferation of rat INS-1E insulinoma cells. J. Ethnopharmacol. 2013, 150, 85–94.
- Ansari, P.; Flatt, P.R.; Harriott, P.; Abdel-Wahab, Y.H.A. Insulin secretory and antidiabetic actions of Heritiera fomes bark together with isolation of active phytomolecules. PLoS ONE 2022, 17, e0264632.
- Hager, R.; Pitsch, J.; Kerbl-Knapp, J.; Neuhauser, C.; Ollinger, N.; Iken, M.; Ranner, J.; Mittermeier-Kleßinger, V.; Dawid, C.; Lanzerstorfer, P.; et al. A High-Content Screen for the Identification of Plant Extracts with Insulin Secretion-Modulating Activity. Pharmaceuticals 2021, 14, 809.
- Kalwat, M.A.; Wichaidit, C.; Nava Garcia, A.Y.; McCoy, M.K.; McGlynn, K.; Hwang, I.H.; MacMillan, J.B.; Posner, B.A.; Cobb, M.H. Insulin promoter-driven Gaussia luciferase-based insulin secretion biosensor assay for discovery of β-cell glucose-sensing pathways. ACS Sens. 2016, 1, 1208–1212.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
1.1K
Entry Collection:
Biopharmaceuticals Technology
Revisions:
2 times
(View History)
Update Date:
01 Aug 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




