Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Colyn Crane-Robinson | -- | 1349 | 2022-07-01 02:59:57 | | | |
| 2 | Catherine Yang | -4 word(s) | 1345 | 2022-07-01 03:02:52 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Crane-Robinson, C. Role of Water in B-Form DNA. Encyclopedia. Available online: https://encyclopedia.pub/entry/24715 (accessed on 07 February 2026).
Crane-Robinson C. Role of Water in B-Form DNA. Encyclopedia. Available at: https://encyclopedia.pub/entry/24715. Accessed February 07, 2026.
Crane-Robinson, Colyn. "Role of Water in B-Form DNA" Encyclopedia, https://encyclopedia.pub/entry/24715 (accessed February 07, 2026).
Crane-Robinson, C. (2022, July 01). Role of Water in B-Form DNA. In Encyclopedia. https://encyclopedia.pub/entry/24715
Crane-Robinson, Colyn. "Role of Water in B-Form DNA." Encyclopedia. Web. 01 July, 2022.
Copy Citation
DNA in the cell is rarely naked but normally protein-bound in nucleosomes. Of special interest is the DNA bound to other factors that control its key functions of transcription, replication, and repair. For these several transactions of DNA, the state of hydration plays an important role in its function, and therefore needs to be defined in as much detail as possible. High-resolution crystallography of short B-form duplexes shows that the mixed polar and apolar surface of the major groove binds water molecules over the broad polar floor of the groove in a sequence-dependent varied manner.
DNA hydration
major groove
minor groove
1. Heat Capacity Changes upon DBD Association with DNA
If the thermodynamic characteristics of protein binding to the two grooves of DNA differ in terms of their unequal states of hydration, this should be reflected in the heat capacity changes induced by protein binding to the two grooves. Heat capacity changes are a good proxy for hydration changes since conformational modifications are too insubstantial to have much influence on the heat capacity, but reductions in hydration have a large effect on the heat capacity change that accompanies its release on DBD binding. Dehydration characteristics on DBD binding to the two grooves of DNA have been studied using the same formalism adopted for protein folding/denaturation. An equation was established for the observed magnitude of ∆Cp for protein folding in terms of the loss of accessible surface area (∆ASA in Å2), both apolar and polar in character [1]:
in which the ∆Cp coefficients are expressed in J K−1 mol−1 (Å2)−1 and ΔASA are in Å2.
ΔCpprotein = −1.79 × ΔASAapolar + 0.98 × ΔASApolar
This equation quantifies the reduction in heat capacity resulting from dehydrating apolar surface and the increase in heat capacity upon dehydrating the polar surface of proteins, for which the former effect dominates to give a substantial overall negative value of ∆Cp. When applied to DNA folding (as detailed above), it explains why ∆Cp is still negative—as for proteins—but lesser in magnitude as a result of the greater polar surface buried.
For a particular DBD/DNA interaction (major or minor groove), this equation can be used to calculate the contribution of protein dehydration to the observed (overall) ∆Cp and then, by difference, obtain the contribution from dehydration of the DNA surface. It can be seen from Figure 1 that the contribution to ∆Cp from the protein components (in orange) is fairly similar in the two grooves, but this is not the case for the component from the dehydrating DNA surface. In the major groove, there is a reduction of about −1 J K−1 mol−1 (Å2)−1 in the heat capacity, but this drops to about −0.3 J K−1 mol−1 (Å2)−1 in the minor groove. To deconvolute the DNA contributions into their polar and apolar components, equations of the above type are established with the two coefficients as unknowns. With several such equations for different DBDs, they can be solved simultaneously for each groove. The resulting data [2] show that the apolar coefficients in the two grooves are similar at about −3 J K−1 mol−1 (Å2)−1, i.e., significantly negative and not so different from that derived for proteins. Regarding the dehydration of the polar surface, the two grooves are very different from each other. In the major groove, the polar coefficient is +0.38 J K−1 mol−1 (Å2)−1, which is positive as with proteins, albeit of lower magnitude than the +0.98 J K−1 mol−1 (Å2)−1 given in Equation (1), in reflection of a less polar state in the major groove. The minor groove is dramatically different: the polar coefficient is +2.67 J K−1 mol−1 (Å2)−1, i.e., positive (as expected), but much more positive than one would predict from observation of the minor groove surface. Such a large positive value can only be a consequence of displacing well-ordered water from the polar groups of the minor groove (N3 of A and O2 of T bases) [2]. It is worth recalling here that the heat capacity of ice is about half that of bulk liquid water.
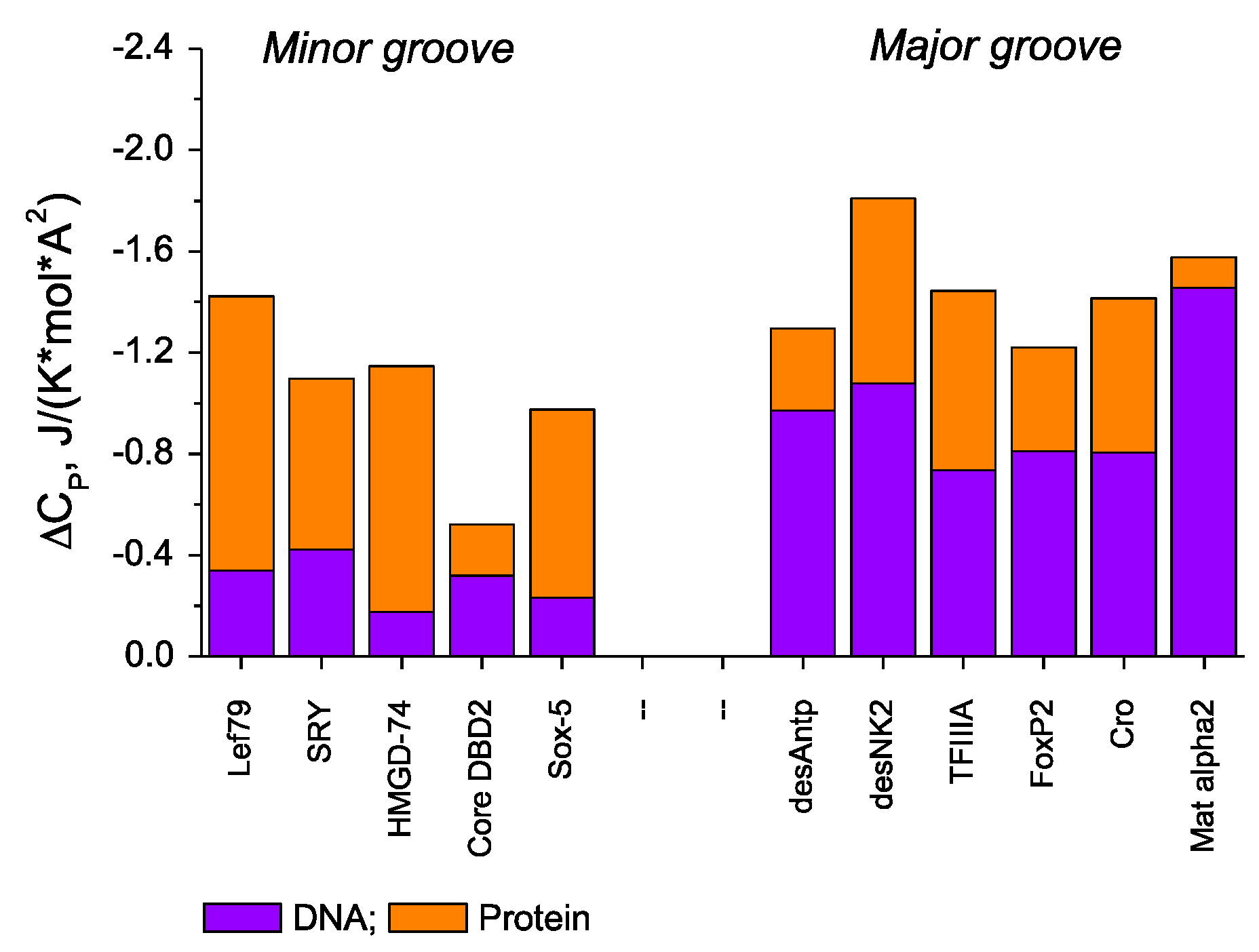
Figure 1. Surface-normalized (i.e., per A2) observed heat capacity changes, ΔCpobs, for binding DBDs to their optimal recognition target sequences. The contribution from protein components (in orange), ΔCpprot, was calculated using Equation (1). The DNA contributions, ΔCpDNA (in blue), were obtained by subtraction from ΔCpobs [2][3].
The several lines of evidence for ordered water in the minor groove make it clear why the overall ∆Cp for melting DNA, while slightly positive, is of much lower magnitude than the large increase in Cp for melting proteins. The very substantial and dominant positive contribution to ∆Cp from the hydration of the hydrophobic surface of polypeptides on melting is opposed by only a small negative component from the hydration of polar groups. However, with DNA, the loss of ordered water from the minor groove on melting generates a very substantial negative effect on ∆Cp—a situation not occurring in proteins [4].
2. DNA Bending
Large bends in DNA are most easily generated by protein binding to the minor groove at AT-rich regions, e.g., by HMG boxes or TATA box binding protein (TBP). Such bending does not appear to demand any extra free energy in that the largest bends (induced by wedge insertion) are found to have the highest affinity: ∆Gnel becomes increasingly negative as the bend angle increases. The characteristic stiffness of the duplex—as seen in its long persistence length of about 45 nm [5], seems to have been eliminated. A reasonable, although partial, explanation for the loss in rigidity is that the stiffness is maintained by the rigid array of water molecules in the minor groove associated with AT pairs. The energetics of DNA bending are discussed in more detail in Refs. [6][7].
3. The Role of Hydration in Enthalpy/Entropy Compensation (EEC)
Modifications to interacting systems frequently lead to compensating alterations in both the enthalpy and entropy of the process, i.e., the Gibbs free energy is barely altered despite large compensating changes occurring to the enthalpy and entropy of the process. This situation is frequently observed when changes are made to ligands that bind to proteins or to DNA. Such enthalpy–entropy compensation (EEC) for a ligand of increased affinity is normally assumed to be a consequence of forming tighter van der Waals contacts to the substrate—contacts that give rise to a more negative enthalpy. However, the additional molecular constraints imposed on both the ligand and the substrate result in a reduction in the conformational entropy, i.e., a more negative ∆S, and this compensates for the more negative enthalpy.
EEC is a widely observed phenomenon, i.e., it appears to be an intrinsic property covering many types of non-covalent systems. In contrast to conformational explanations of EEC, changes in the solvation of a system can also contribute to EEC and frequently dominate the measured enthalpy/entropy components. The contribution to EEC from modified hydration is frequently ignored, and elaborate hypothetical structural explanations offered for the observed compensating enthalpy/entropy changes.
A particularly revealing example of solvation-based enthalpy–entropy compensation is the yeast bZIP DBD from GCN4, binding as a crosslinked homodimer in a scissors grip to target DNA elements of a slightly different sequence, AP-1 and ATF/CREB, Refs. [8][9]. These targets differ in sequence by just one base pair. However, the crystal structures of the complexes with the two DNAs show them to be very similar [10][11]: the two α-helix backbones that contact the DNA major groove overlap each other with an RMSD of only 1.3 Å.
The most reasonable explanation for these very large discrepancies in the entropies and enthalpies of forming the two very similar GCN4 complexes is differences in the number of incorporated ordered water molecules. If we approximate the immobilization/release of water molecules as similar to that of freezing/melting water, it follows that the AP-1 complex has seven or eight more incorporated water molecules than the ATF/CREB complex [8]. It is worth recalling that the binding/release of dynamically constrained water from macromolecular systems is intrinsically compensatory: if such water has an ice-like structure and the temperature is 273 K, there would be no consequent change in the Gibbs energy despite large changes in the enthalpy and entropy. Solvation changes, therefore, represent the principal energetic basis of enthalpy–entropy compensation [9].
References
- Makhatadze, G.I.; Privalov, P.L. Energetics of protein structure. Adv. Prot. Chem. 1995, 47, 307–425.
- Dragan, A.I.; Read, C.M.; Crane-Robinson, C. Hydration differences between the major and minor grooves of DNA revealed from heat capacity measurements. Eur. Biophys. J. 2019, 48, 131–138.
- Privalov, P.L.; Crane-Robinson, C. Role of water in the formation of macromolecular structures. Eur. Biophys. J. 2017, 46, 203–224.
- Dragan, A.I.; Privalov, P.L.; Crane-Robinson, C. Thermodynamics of DNA: Heat capacity changes on duplex unfolding. Eur. Biophys. J. 2019, 48, 773–779.
- Taylor, W.H.; Hagerman, P.J. Application of the method of phage T4 DNA ligase-catalyzed ring-closure to the study of DNA structure. J. Mol. Biol. 1990, 212, 363–376.
- Privalov, P.L.; Crane-Robinson, C. Translational Entropy and DNA Duplex Stability. Biophys. J. 2018, 114, 15–20.
- Privalov, P.L.; Dragan, A.I.; Crane-Robinson, C. The cost of DNA bending. Trends Biochem. Sci. 2009, 34, 464–470.
- Dragan, A.I.; Frank, L.; Liu, Y.; Makeyeva, E.N.; Crane-Robinson, C.; Privalov, P.L. Thermodynamic Signature of GCN4-bZIP Binding to DNA Indicates the Role of Water in Discriminating Between the AP-1 and ATF/CREB Sites. J. Mol. Biol. 2004, 343, 865–878.
- Dragan, A.I.; Read, C.M.; Crane-Robinson, C. Enthalpy–entropy compensation: The role of solvation. Eur. Biophys. J. 2017, 46, 301–308.
- Ellenberger, T.E.; Brandl, C.J.; Struhl, K.; Harrison, S.C. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted alpha Helices: Crystal structure of the protein-DNA complex. Cell 1992, 71, 1223–1237.
- Keller, W.; Konig, P.; Richmond, T.J. Crystal structure of a bZIP/DNA complex at 2.2 Å: Determinants of DNA specific recognition. J. Mol. Biol. 1995, 254, 657–667.
More
Information
Subjects:
Chemistry, Inorganic & Nuclear
Contributor
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
730
Revisions:
2 times
(View History)
Update Date:
01 Jul 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




