| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Jose Luis Rosa | + 1040 word(s) | 1040 | 2020-10-05 02:31:32 | | | |
| 2 | Bruce Ren | Meta information modification | 1040 | 2020-10-09 05:52:52 | | | | |
| 3 | Bruce Ren | Meta information modification | 1040 | 2020-10-21 10:00:16 | | |
Video Upload Options
HERC proteins are ubiquitin E3 ligases of the HECT family. The HERC subfamily is composed of six members classified by size into large (HERC1 and HERC2) and small (HERC3–HERC6). HERC family ubiquitin ligases regulate important cellular processes, such as neurodevelopment, DNA damage response, cell proliferation, cell migration, and immune responses. Accumulating evidence also shows that this family plays critical roles in cancer. In this review, we provide an integrated view of the role of these ligases in cancer, highlighting their bivalent functions as either oncogenes or tumor suppressors, depending on the tumor type.
1. Introduction
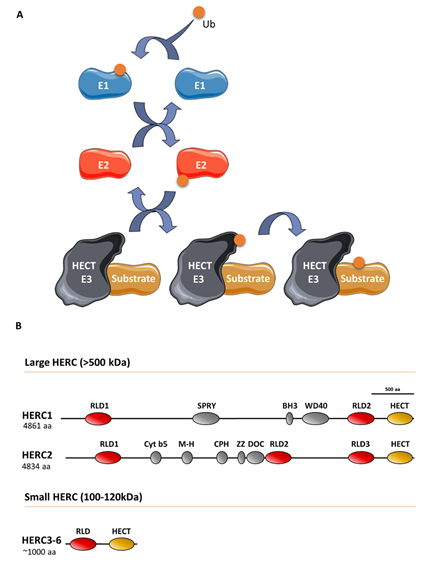
Ubiquitin E3 ligases take part in protein ubiquitylation. These enzymes catalyze the last step of a cascade where ubiquitin is initially incorporated to a ubiquitin-activating enzyme (E1), which in turn is transferred to a ubiquitin-conjugating enzyme (E2), and finally, to a target protein through a process defined by a ubiquitin E3 ligase that interacts with the substrate protein (Figure 1). The ubiquitin-like proteins SUMO, NEDD8, and ISG15 are also covalently attached to the target protein via an E1/E2/E3 cascade. Specifically, the E3 ligases can be classified into three groups, of which one is homologous to the E6AP carboxyl terminus (HECT) protein. All HECT ligases have a catalytic domain in their carboxyl terminus that contains a conserved cysteine residue that is involved in forming a transiently thioester bond to ubiquitin before transferring it to the lysine residue of the substrate protein (Figure 1) [1]. HECT ligases containing one or more regulator of chromosome condensation 1 (RCC1)-like domains in their amino-terminal domain form a HERC subgroup [2]. HERC1 and HERC2 are the largest HECT ligases, having molecular weights exceeding 500 kDa, and constitute the large HERC protein subfamily [3]. By contrast, HERC3 to HERC6 have molecular weights around 100–120 kDa and constitute the small HERC protein subfamily. Despite the structural similarity between large and small HERC proteins (Figure 1), they are evolutionarily very distant. In fact, they are the result of convergence phenomena rather than being phylogenetic paralogs [3][4][5]. Moreover, the small HERC proteins HERC5 and HERC6 may also function as ISG15 E3 ligases [6][7].
Figure 1. The ubiquitin-conjugating system in HERC E3 ligases: (A) Ubiquitin (Ub) is conjugated to a target substrate via a cascade that comprises an E1 activating enzyme, an E2 conjugating enzyme, and an E3 ligase enzyme. The HERC proteins belong to the HECT family of E3 ligases, which form a thioester bond with Ub via a conserved cysteine residue. Once formed, Ub is transferred to the substrate’s lysine residue (see text for details). Ub-like proteins, such as ISG15, are also covalently attached to the substrate protein via an E1/E2/E3 cascade; (B) structural features of large and small HERC proteins are also shown. HERC5 and HERC6 may also function as ISG15 E3 ligases.
2. The Role of HERCs in Cancer
HERCs play roles in a wide range of cellular functions, including neurodevelopment, cell response to replication stress and DNA damage, cell proliferation, cell migration, and immune responses. As such, mutations in HERCs are associated with severe pathologies [8], with a notable impact in cancer. An extensive list of the different cancers associated with the specific large and small HERCs is provided in Table 1.
Table 1. Cancers associated with HERCs and related molecular mechanisms.
|
Genes |
Associated Cancers |
Related Molecular Mechanisms |
Reference |
|
HERC1 |
Acute promyelocytic leukemia |
HERC1-PML genomic fusion |
[9] |
|
Acute Myeloid Leukemia |
HERC1 mutations |
[10] |
|
|
Acute lymphoblastic leukemia |
Decreased MSH2 protein levels and HERC1 deletions |
[11] |
|
|
Adult T-cell acute lymphoblastic leukemia |
HERC1 mutations |
[12] |
|
|
T-cell prolymphocytic leukemia |
HERC1 mutations |
[13] |
|
|
Non-melanoma skin cancer |
Enhanced BAK protein degradation |
[14] |
|
|
Pulmonary sclerosing pneumocytoma |
HERC1 mutations |
[15] |
|
|
Invasive lobular breast cancer |
HERC1 mutations |
[16] |
|
|
Metastatic triple-negative breast cancer |
HERC1 mutations |
[17] |
|
|
Sporadic colorectal cancer |
Decreased MSH2 protein levels and HERC1 deletions |
[11] |
|
|
Osteosarcoma |
Negative correlation of SOX18 overexpression and HERC1 mRNA levels |
[18] |
|
|
HERC2 |
Pheochromocytoma and paraganglioma |
HERC2 mutations |
[19] |
|
T-cell prolymphocytic leukemia |
HERC2 mutations |
[13] |
|
|
Cutaneous melanoma |
SNPs in HERC2 gene increase susceptibility |
||
|
Gene-gene interactions between HERC2 gene and IL31RA and DDX4 genes |
[24] |
||
|
Epistatic effects between HERC2 and VDR genes |
[25] |
||
|
Cutaneous squamous cell carcinoma |
SNPs in HERC2 gene impact on time to develop the tumor in organ transplant recipients |
[26] |
|
|
Uveal melanoma |
SNPs in HERC2 gene increase susceptibility |
[27] |
|
|
Non-small-cell lung cancer |
Worse prognosis in patients expressing high HERC2 mRNA levels |
[28] |
|
|
Breast cancer |
Enhanced BRCA1 degradation |
||
|
Gastric and colorectal carcinomas |
HERC2 mutations |
[31] |
|
|
Osteosarcoma |
Negative correlation of SOX18 overexpression and HERC2 mRNA levels |
[18] |
|
|
HERC3 |
Glioblastoma |
Degradation of SMAD7 and activation of the TGFβ signaling |
[32] |
|
Gastric and colorectal carcinomas |
HERC3 mutations |
[31] |
|
|
Osteosarcoma |
Negative correlation of SOX18 overexpression and HERC3 mRNA levels |
[18] |
|
|
HERC4 |
Multiple myeloma |
Decreased c-Maf degradation |
[33] |
|
Lung cancer |
HERC4 overexpression |
[34] |
|
|
Non-small cell lung cancer |
Increased Smo protein stability and Hh pathway activation |
||
|
Breast cancer |
HERC4 upregulation |
[37] |
|
|
Decreased expression of miRNAs targeting HERC4 expression and enhanced LATS1 degradation |
[38] |
||
|
Hepatocellular carcinoma |
HERC4 overexpression |
[39] |
|
|
Osteosarcoma |
Negative correlation of SOX18 overexpression and HERC4 mRNA levels |
[18] |
|
|
HERC5 |
Pediatric germ cell tumors |
Chromosome copy number variations (CNVs) at a region encompassing HERC5 gene |
[40] |
|
Glioblastoma |
HERC5 upregulation |
[41] |
|
|
Acute myeloid leukemia |
HERC5 downregulation |
[42] |
|
|
Oropharyngeal cancer |
HERC5 gene expression is associated with overall survival |
[43] |
|
|
Non-small cell lung cancer |
HERC5 promoter hypermethylation |
[44] |
|
|
Breast cancer |
HERC5 upregulation |
[45] |
|
|
Hepatocellular carcinoma |
Negative correlation of CCL20 overexpression and HERC5 mRNA levels |
[46] |
|
|
Reduced p53, p21 and Bax/Bcl-2 pathway activation |
[47] |
||
|
Ovarian cancer |
HERC5 upregulation is associated with drug resistance |
||
|
Osteosarcoma |
Negative correlation of SOX18 overexpression and HERC5 mRNA levels |
[18] |
|
|
HERC6 |
Osteosarcoma |
Negative correlation of SOX18 overexpression and HERC6 mRNA levels |
[18] |
Mutations in large HERCs have been found in leukemia and breast cancer . Frameshift mutations in HERC2 have been found in both gastric and colorectal carcinomas with microsatellite instability. The HERC2 locus has also been associated with both cutaneous melanoma and uveal melanoma, whereas the HERC1 locus has been found to be mutated in non-melanoma skin cancer. Higher expression levels of HERCs are associated with better patient prognosis in kidney, head and neck, and pancreatic cancers when HERC1 expression levels are elevated, and in patients with renal cancer when HERC2 expression levels are elevated [50]. By contrast, the expression levels of HERC2 have been found to negatively correlate with patient survival in non-small-cell lung cancer . In osteosarcoma, upregulation of the HERC2-binding protein SOX18 enhances cell proliferation, and it correlates with a reduction in both large and small HERC mRNA levels (Table 1) .
References
- Ciechanover, A. The unravelling of the ubiquitin system. Nat. Rev. Mol. Cell Biol. 2015, 16, 322–324.
- Garcia-Gonzalo, F.R.; Rosa, J.L. The HERC proteins: Functional and evolutionary insights. Cell. Mol. Life Sci. 2005, 62, 1826–1838.
- García-Cano, J.; Martinez-Martinez, A.; Sala-Gaston, J.; Pedrazza, L.; Rosa, J.L. HERCing: Structural and Functional Relevance of the Large HERC Ubiquitin Ligases. Front. Physiol. 2019, 10, 1014.
- Hadjebi, O.; Casas-Terradellas, E.; Garcia-Gonzalo, F.R.; Rosa, J.L. The RCC1 superfamily: From genes, to function, to disease. Biochim. Biophys. Acta Mol. Cell Res. 2008, 1783, 1467–1479.
- Marin, I. Animal HECT ubiquitin ligases: Evolution and functional implications. BMC Evol. Biol. 2010, 10, 56.
- Sánchez-Tena, S.; Cubillos-Rojas, M.; Schneider, T.; Rosa, J.L. Functional and pathological relevance of HERC family proteins: A decade later. Cell. Mol. Life Sci. 2016, 73, 1955–1968.
- Villarroya-Beltri, C.; Guerra, S.; Sánchez-Madrid, F. ISGylation—A key to lock the cell gates for preventing the spread of threats. J. Cell Sci. 2017, 130, 2961–2969.
- Mao, X.; Sethi, G.; Zhang, Z.; Wang, Q. The Emerging Roles of the HERC Ubiquitin Ligases in Cancer. Curr. Pharm. Des. 2018, 24, 1676–1681.
- Walz, C.; Grimwade, D.; Saussele, S.; Lengfelder, E.; Haferlach, C.; Schnittger, S.; Lafage-Pochitaloff, M.; Hochhaus, A.; Cross, N.C.P.; Reiter, A. Atypical mRNA fusions in PML-RARA positive, RARA-PML negative acute promyelocytic leukemia. Genes Chromosom. Cancer 2010, 49, 471–479.
- Opatz, S.; Bamopoulos, S.A.; Metzeler, K.H.; Herold, T.; Ksienzyk, B.; Bräundl, K.; Tschuri, S.; Vosberg, S.; Konstandin, N.P.; Wang, C.; et al. The clinical mutatome of core binding factor leukemia. Leukemia 2020, 34, 1553–1562.
- Diouf, B.; Pei, D.; Fan, Y.; Cheng, C.; Krynetskiy, E.Y.; Geng, H.; Chen, S.; Thierfelder, W.E.; Mullighan, C.G.; Downing, J.R.; et al. Somatic deletions of genes regulating MSH2 protein stability cause DNA mismatch repair deficiency and drug resistance in human leukemia cells. Nat. Med. 2012, 17, 1298–1303.
- Neumann, M.; Vosberg, S.; Schlee, C.; Heesch, S.; Schwartz, S.; Gökbuget, N.; Hoelzer, D.; Graf, A.; Krebs, S.; Bartram, I.; et al. Mutational spectrum of adult T-ALL. Oncotarget 2015, 6, 2754–2766.
- Johansson, P.; Klein-Hitpass, L.; Choidas, A.; Habenberger, P.; Mahboubi, B.; Kim, B.; Bergmann, A.; Scholtysik, R.; Brauser, M.; Lollies, A.; et al. SAMHD1 is recurrently mutated in T-cell prolymphocytic leukemia. Blood Cancer J. 2018, 8, 11.
- Holloway, A.; Simmonds, M.; Azad, A.; Fox, J.L.; Storey, A. Resistance to UV-induced apoptosis by β-HPV5 E6 involves targeting of activated BAK for proteolysis by recruitment of the HERC1 ubiquitin ligase. Int. J. Cancer 2015, 136, 2831–2843.
- Fan, X.; Lin, L.; Wang, J.; Wang, Y.; Feng, A.; Nie, L.; Wu, H.; Meng, F.; Xu, H. Genome profile in a extremely rare case of pulmonary sclerosing pneumocytoma presenting with diffusely-scattered nodules in the right lung. Cancer Biol. Ther. 2018, 19, 13–19.
- Ping, Z.; Siegal, G.P.; Harada, S.; Eltoum, I.E.; Youssef, M.; Shen, T.; He, J.; Huang, Y.; Chen, D.; Li, Y.; et al. ERBB2 mutation is associated with a worse prognosis in patients with CDH1 altered invasive lobular cancer of the breast. Oncotarget 2016, 7, 80655–80663.
- Craig, D.W.; O’Shaughnessy, J.A.; Kiefer, J.A.; Aldrich, J.; Sinari, S.; Moses, T.M.; Wong, S.; Dinh, J.; Christoforides, A.; Blum, J.L.; et al. Genome and transcriptome sequencing in prospective metastatic triple-negative breast cancer uncovers therapeutic vulnerabilities. Mol. Cancer Ther. 2013, 12, 104–116.
- Zhu, D.; Yang, D.; Li, X.; Feng, F. Heterogeneous expression and biological function of SOX18 in osteosaroma. J. Cell. Biochem. 2018, 119, 4184–4192.
- Suh, Y.J.; Choe, J.Y.; Park, H.J. Malignancy in Pheochromocytoma or Paraganglioma: Integrative Analysis of 176 Cases in TCGA. Endocr. Pathol. 2017, 28, 159–164.
- Ibarrola-Villava, M.; Fernandez, L.P.; Pita, G.; Bravo, J.; Floristan, U.; Sendagorta, E.; Feito, M.; Avilés, J.A.; Martin-Gonzalez, M.; Lázaro, P.; et al. Genetic analysis of three important genes in pigmentation and melanoma susceptibility: CDKN2A, MC1R and HERC2/OCA2. Exp. Dermatol. 2010, 19, 836–844.
- Amos, C.I.; Wang, L.E.; Lee, J.E.; Gershenwald, J.E.; Chen, W.V.; Fang, S.; Kosoy, R.; Zhang, M.; Qureshi, A.A.; Vattathil, S.; et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum. Mol. Genet. 2011, 20, 5012–5023.
- Fesenko, D.O.; Chudinov, A.V.; Surzhikov, S.A.; Zasedatelev, A.S. Biochip-Based Genotyping Assay for Detection of Polymorphisms in Pigmentation Genes Associated with Cutaneous Melanoma. Genet. Test. Mol. Biomarkers 2016, 20, 208–212.
- Laino, A.M.; Berry, E.G.; Jagirdar, K.; Lee, K.J.; Duffy, D.L.; Soyer, H.P.; Sturm, R.A. Iris pigmented lesions as a marker of cutaneous melanoma risk: An Australian case–control study. Br. J. Dermatol. 2018, 178, 1119–1127.
- Xiao, F.; Ma, J.; Cai, G.; Fang, S.; Lee, J.E.; Wei, Q.; Amos, C.I. Natural and orthogonal model for estimating gene-gene interactions applied to cutaneous melanoma. Hum. Genet. 2014, 133, 559–574.
- Kosiniak-Kamysz, A.; Marczakiewicz-Lustig, A.; Marcińska, M.; Skowron, M.; Wojas-Pelc, A.; Pośpiech, E.; Branicki, W. Increased risk of developing cutaneous malignant melanoma is associated with variation in pigmentation genes and VDR, and may involve epistatic effects. Melanoma Res. 2014, 24, 388–396.
- Wei, L.; Allain, D.C.; Bernhardt, M.N.; Gillespie, J.L.; Peters, S.B.; Iwenofu, O.H.; Nelson, H.H.; Arron, S.T.; Toland, A.E. Variants at the OCA2/HERC2 locus affect time to first cutaneous squamous cell carcinoma in solid organ transplant recipients collected using two different study designs. Br. J. Dermatol. 2017, 177, 1066–1073.
- Ferguson, R.; Vogelsang, M.; Ucisik-Akkaya, E.; Rai, K.; Pilarski, R.; Martinez, C.N.; Rendleman, J.; Kazlow, E.; Nagdimov, K.; Osman, I.; et al. Genetic markers of pigmentation are novel risk loci for uveal melanoma. Sci. Rep. 2016, 6, 31191.
- Bonanno, L.; Costa, C.; Majem, M.; Sanchez, J.J.; Rodriguez, I.; Gimenez-Capitan, A.; Molina-Vila, M.A.; Vergnenegre, A.; Massuti, B.; Favaretto, A.; et al. Combinatory effect of BRCA1 and HERC2 expression on outcome in advanced non-small-cell lung cancer. BMC Cancer 2016, 16, 312.
- Wu, W.; Sato, K.; Koike, A.; Nishikawa, H.; Koizumi, H.; Venkitaraman, A.R.; Ohta, T. HERC2 is an E3 ligase that targets BRCA1 for degradation. Cancer Res. 2010, 70, 6384–6392.
- Peng, Y.; Dai, H.; Wang, E.; Lin, C.C.J.; Mo, W.; Peng, G.; Lin, S.Y. TUSC4 functions as a tumor suppressor by regulating BRCA1 stability. Cancer Res. 2015, 75, 378–386.
- Yoo, N.J.; Park, S.W.; Lee, S.H. Frameshift mutations of ubiquitinationrelated genes HERC2, HERC3, TRIP12, UBE2Q1 and UBE4B in gastric and colorectal carcinomas with microsatellite instability. Pathology 2011, 43, 753–755.
- Li, H.; Li, J.; Chen, L.; Qi, S.; Yu, S.; Weng, Z.; Hu, Z.; Zhou, Q.; Xin, Z.; Shi, L.; et al. HERC3-Mediated SMAD7 ubiquitination degradation promotes autophagy-induced EMT and chemoresistance in Glioblastoma. Clin. Cancer Res. 2019, 25, 3602–3616.
- Zhang, Z.; Tong, J.; Tang, X.; Juan, J.; Cao, B.; Hurren, R.; Chen, G.; Taylor, P.; Xu, X.; Shi, C.X.; et al. The ubiquitin ligase HERC4 mediates c-Maf ubiquitination and delays the growth of multiple myeloma xenografts in nude mice. Blood 2016, 127, 1676–1686.
- Zeng, W.L.; Chen, Y.W.; Zhou, H.; Zhou, J.Y.; Wei, M.; Shi, R. Expression of HERC4 in lung cancer and its correlation with clinicopathological parameters. Asian Pac. J. Cancer Prev. 2015, 16, 513–517.
- Jiang, W.; Yao, X.; Shan, Z.; Li, W.; Gao, Y.; Zhang, Q. E3 ligase Herc4 regulates Hedgehog signalling through promoting Smoothened degradation. J. Mol. Cell Biol. 2019, 11, 791–803.
- Sun, X.; Sun, B.; Cui, M.; Zhou, Z. HERC4 exerts an anti-tumor role through destabilizing the oncoprotein Smo. Biochem. Biophys. Res. Commun. 2019, 513, 1013–1018.
- Zhou, H.; Shi, R.; Wei, M.; Zheng, W.L.; Zhou, J.Y.; Ma, W.L. The expression and clinical significance of HERC4 in breast cancer. Cancer Cell Int. 2013, 13, 113.
- Xu, Y.; Ji, K.; Wu, M.; Hao, B.; Yao, K. tai; Xu, Y. A miRNA-HERC4 pathway promotes breast tumorigenesis by inactivating tumor suppressor LATS1. Protein Cell 2019, 10, 595–605.
- Zheng, Y.; Li, J.; Pan, C.; Zhou, G.; Zhuge, L.; Jin, L.; Fang, P. HERC4 Is Overexpressed in Hepatocellular Carcinoma and Contributes to the Proliferation and Migration of Hepatocellular Carcinoma Cells. DNA Cell Biol. 2017, 36, 490–500.
- Wang, H.W.; Wu, Y.H.; Hsieh, J.Y.; Liang, M.L.; Chao, M.E.; Liu, D.J.; Hsu, M.T.; Wong, T.T. Pediatric primary central nervous system germ cell tumors of different prognosis groups show characteristic miRNome traits and chromosome copy number variations. BMC Genom. 2010, 11, 132.
- Bhargava, S.; Patil, V.; Mahalingam, K.; Somasundaram, K. Elucidation of the genetic and epigenetic landscape alterations in RNA binding proteins in glioblastoma. Oncotarget 2017, 8, 16650–16668.
- Niu, P.; Yao, B.; Wei, L.; Zhu, H.; Fang, C.; Zhao, Y. Construction of prognostic risk prediction model based on high-throughput sequencing expression profile data in childhood acute myeloid leukemia. Blood Cells Mol. Dis. 2019, 77, 43–50.
- Reddy, R.B.; Khora, S.S.; Suresh, A. Molecular prognosticators in clinically and pathologically distinct cohorts of head and neck squamous cell carcinoma—A meta-analysis approach. PLoS ONE 2019, 14, e0218989.
- Wrage, M.; Hagmann, W.; Kemming, D.; Uzunoglu, F.G.; Riethdorf, S.; Effenberger, K.; Westphal, M.; Lamszus, K.; Kim, S.Z.; Becker, N.; et al. Identification of HERC5 and its potential role in NSCLC progression. Int. J. Cancer 2015, 136, 2264–2272.
- Tang, J.; Yang, Q.; Cui, Q.; Zhang, D.; Kong, D.; Liao, X.; Ren, J.; Gong, Y.; Wu, G. Weighted gene correlation network analysis identifies RSAD2, HERC5, and CCL8 as prognostic candidates for breast cancer. J. Cell. Physiol. 2020, 235, 394–407.
- Xue, F.; Higgs, B.W.; Huang, J.; Morehouse, C.; Zhu, W.; Yao, X.; Brohawn, P.; Xiao, Z.; Sebastian, Y.; Liu, Z.; et al. HERC5 is a prognostic biomarker for post-liver transplant recurrent human hepatocellular carcinoma. J. Transl. Med. 2015 13, 379.
- Wang, Y.; Ding, Q.; Xu, T.; Li, C. yao; Zhou, D. dan; Zhang, L. HZ-6d targeted HERC5 to regulate p53 ISGylation in human hepatocellular carcinoma. Toxicol. Appl. Pharmacol. 2017, 334, 180–191.
- Januchowski, R.; Sterzyńska, K.; Zawierucha, P.; Ruciński, M.; Świerczewska, M.; Partyka, M.; Bednarek-Rajewska, K.; Brazert, M.; Nowicki, M.; Zabel, M.; et al. Microarray-based detection and expression analysis of new genes associated with drug resistance in ovarian cancer cell lines. Oncotarget 2017, 8, 49944–49958.
- Swierczewska, M.; Klejewski, A.; Wojtowicz, K.; Brazert, M.; Izycki, D.; Nowicki, M.; Zabel, M.; Januchowski, R. New and old genes associated with primary and established responses to cisplatin and topotecan treatment in ovarian cancer cell lines. Molecules 2017, 22, 1717.
- Schneider, T.; Martinez-Martinez, A.; Cubillos-Rojas, M.; Bartrons, R.; Ventura, F.; Rosa, J.L. Large HERCS function as tumor suppressors. Front. Oncol. 2019, 9, 524.