
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Akhila Ajith | -- | 4970 | 2022-05-19 09:09:50 | | | |
| 2 | Vivi Li | + 38 word(s) | 5008 | 2022-05-20 04:07:40 | | |
Video Upload Options
The detection of chemical species and understanding their respective localisations in tissues have important implications in plant science. The conventional methods for imaging spatial localisation of chemical species are often restricted by the number of species that can be identified and is mostly done in a targeted manner. Mass spectrometry imaging combines the ability of traditional mass spectrometry to detect numerous chemical species in a sample with their spatial localisation information by analysing the specimen in a 2D manner. An insight into the spatial localisations of different chemicals in the plant system can be instrumental in understanding the movement and localisation of plant biochemicals and xenobiotics as well as their responses to different stress and metabolic pathways.
1. Introduction
2. Mass Spectrometry Imaging of Plant Vegetative Parts
2.1. Endogenous Compounds
2.1.1. MALDI Imaging Studies
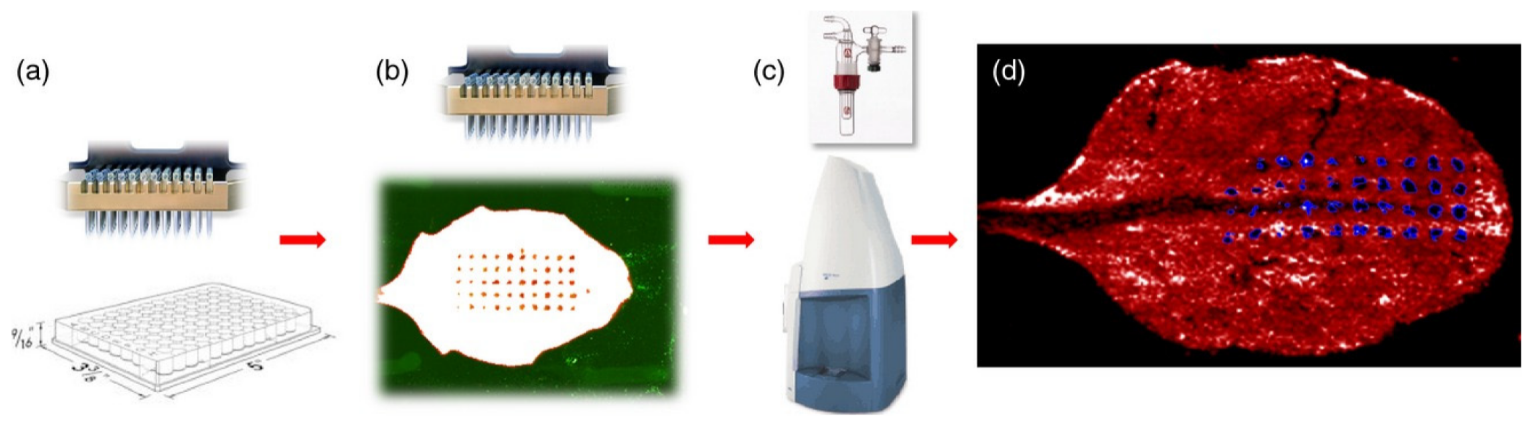
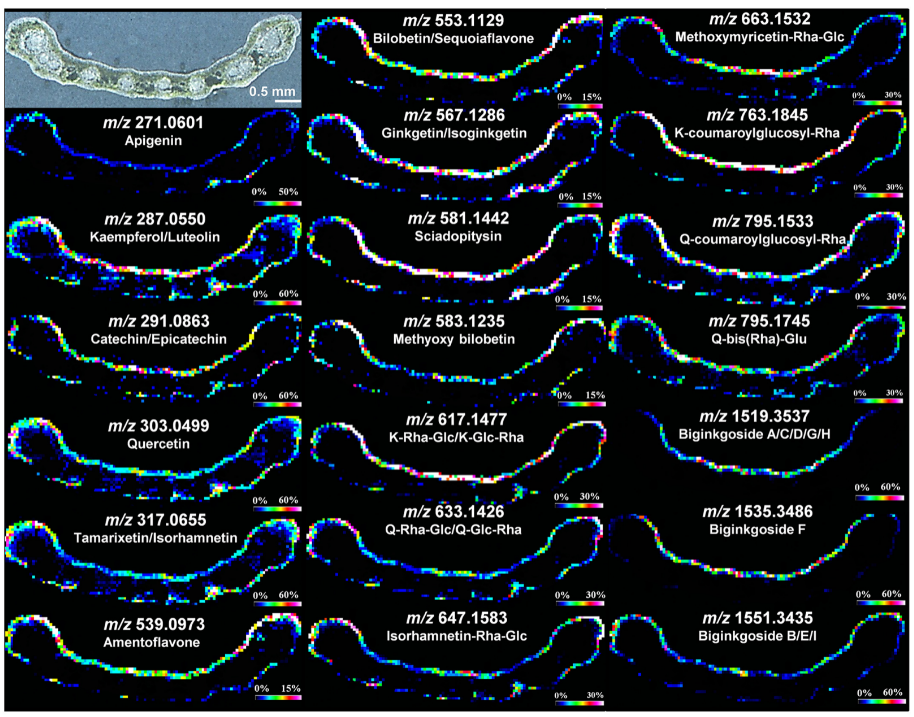
2.1.2. SIMS Imaging
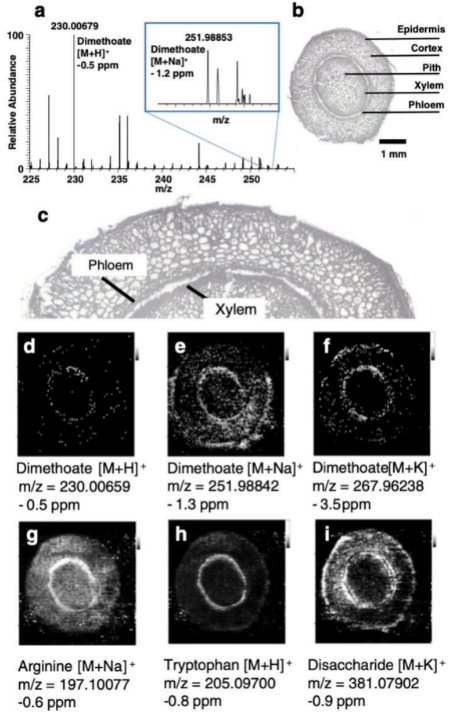
2.2. Crop-Protection Products
| MSI Technique | Plant Species | Agrochemical | Type of Uptake | Reference |
|---|---|---|---|---|
| MALDI | Soy | Mesotrione (Herbicide) Azoxystrobin (Fungicide) |
Leaf and root | [20] |
| MALDI | Sunflower | Nicosulfron (Herbicide) | Leaf and root | [67] |
| MALDI | Wheat | Epoxiconazole Azoxystrobin Pyraclostrobin (All fungicides) |
Leaf | [68] |
| MALDI | Tomato | Metalaxyl (Fungicide) | Root | [69] |
| DESI | Cotoneaster horizontalis Kalanhoe blossfeldiana |
Rapeseed oil and pyrethrins Imidacloprid and Methiocarb Dimethoate (All insecticides) |
Leaf Leaf Root |
[70] |
| LA-APCI | Tomato | Isotianil (Fungicide) | Leaf | [71] |
| Nano-PALDI | Flowering cabbage | Chlorantraniliprole (Insecticide) |
Leaf | [72] |
References
- Hu, W.; Han, Y.; Sheng, Y.; Wang, Y.; Pan, Q.; Nie, H. Mass spectrometry imaging for direct visualizetion of components in plants tissues. J. Sep. Sci. 2021, 44, 3462–3476.
- Sulpice, R.; McKeown, P.C. Moving Toward a Comprehensive Map of Central Plant Metabolism. Annu. Rev. Plant Biol. 2015, 66, 187–210.
- Salem, M.A.; de Souza, L.P.; Serag, A.; Fernie, A.R.; Farag, M.A.; Ezzat, S.M.; Alseekh, S. Metabolomics in the Context of Plant Natural Products Research: From Sample Preparation to Metabolite Analysis. Metabolites 2020, 10, 37.
- Boughton, B.A.; Thinagaran, D.; Sarabia, D.; Bacic, A.; Roessner, U. Mass spectrometry imaging for plant biology: A review. Phytochem. Rev. 2015, 15, 445–488.
- Bhandari, D.R.; Wang, Q.; Friedt, W.; Spengler, B.; Gottwald, S.; Römpp, A. High resolution mass spectrometry imaging of plant tissues: Towards a plant metabolite atlas. Analyst 2015, 140, 7696–7709.
- Rubio, G.; Sorgonà, A.; Lynch, J.P. Spatial mapping of phosphorus influx in bean root systems using digital autoradiography. J. Exp. Bot. 2004, 55, 2269–2280.
- Jacobsen, S.C.; Speth, N.R.; Xiong, M.; Herth, M.M.; Kristensen, J.L.; Palner, M.; Janfelt, C. Desorption Electrospray Ionization Mass Spectrometry Imaging of Cimbi-36, a 5-HT2A Receptor Agonist, with Direct Comparison to Autoradiography and Positron Emission Tomography. Mol. Imaging Biol. 2021, 23, 676–685.
- Boaru, S.G.; Merle, U.; Uerlings, R.; Zimmermann, A.; Flechtenmacher, C.; Willheim, C.; Eder, E.; Ferenci, P.; Stremmel, W.; Weiskirchen, R. Laser ablation inductively coupled plasma mass spectrometry imaging of metals in experimental and clinical Wilson’s disease. J. Cell. Mol. Med. 2015, 19, 806–814.
- Ifa, D.R.; Eberlin, L.S. Ambient Ionization Mass Spectrometry for Cancer Diagnosis and Surgical Margin Evaluation. Clin. Chem. 2016, 62, 111–123.
- Touboul, D.; Roy, S.; Germain, D.P.; Chaminade, P.; Brunelle, A.; Laprévote, O. MALDI-TOF and cluster-TOF-SIMS imaging of Fabry disease biomarkers. Int. J. Mass Spectrom. 2007, 260, 158–165.
- Yeh, K.; Burr, W.S.; Stock, N.L.; Stotesbury, T. Preliminary analysis of latent fingerprints recovered from underneath bloodstains using matrix-assisted laser de-sorption/ionization fourier-transform ion cyclotron resonance mass spectrometry imaging (MALDI FT-ICR MSI). Forensic Chem. 2020, 20, 100274.
- Yoshimura, Y.; Zaima, N.; Yukihiro, Y. Application of Mass Spectrometry Imaging for Visualizing Food Components. Foods 2020, 9, 575.
- Morisasa, M.; Sato, T.; Kimura, K.; Mori, T.; Goto-Inoue, N. Application of Matrix-Assisted Laser Desorption/Ionization Mass Spectrometry Imaging for Food Analysis. Foods 2019, 8, 633.
- Nielen, M.W.; van Beek, T.A. Macroscopic and microscopic spatially-resolved analysis of food contaminants and constituents using laser-ablation electrospray ionization mass spectrometry imaging. Anal. Bioanal. Chem. 2014, 406, 6805–6815.
- Hickert, S.; Cramer, B.; Letzel, M.C.; Humpf, H.-U. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry imaging of ochratoxin A and fumonisins in mold-infected food. Rapid Commun. Mass Spectrom. 2016, 30, 2508–2516.
- Wang, T.-T.; Ying, G.-G.; Shi, W.-J.; Zhao, J.-L.; Liu, Y.-S.; Chen, J.; Ma, D.-D.; Xiong, Q. Uptake and Translocation of Perfluorooctanoic Acid (PFOA) and Perfluorooctanesulfonic Acid (PFOS) by Wetland Plants: Tissue- and Cell-Level Distribution Visualization with Desorption Electrospray Ionization Mass Spectrometry (DESI-MS) and Transmission Electron Microscopy Equipped with Energy-Dispersive Spectroscopy (TEM-EDS). Environ. Sci. Technol. 2020, 54, 6009–6020.
- Villette, C.; Maurer, L.; Wanko, A.; Heintz, D. Xenobiotics metabolization in Salix alba leaves uncovered by mass spectrometry imaging. Metabolomics 2019, 15, 122.
- Ye, H.; Gemperline, E.; Li, L. A vision for better health: Mass spectrometry imaging for clinical diagnostics. Clin. Chim. Acta 2012, 420, 11–22.
- Porta Siegel, T.; Hamm, G.; Bunch, J.; Cappell, J.; Fletcher, J.S.; Schwamborn, K. Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol. Imaging Biol. 2018, 20, 888–901.
- Mullen, A.K.; Clench, M.R.; Crosland, S.; Sharples, K.R. Determination of agrochemical compounds in soya plants by imaging matrix-assisted laser desorption/ionisation mass spectrometry. Rapid Commun. Mass Spectrom. 2005, 19, 2507–2516.
- Ford, M.A.; Blackwell, R.D.; Parker, M.L.; Austin, R.B. Associations between Stem Solidity, Soluble Carbohydrate Accumulation and Other Characters in Wheat. Ann. Bot. 1979, 44, 731–738.
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 2006, 57, 303–333.
- Shroff, R.; Vergara, F.; Muck, A.; Svatoš, A.; Gershenzon, J. Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. USA 2008, 105, 6196–6201.
- Shroff, R.; Schramm, K.; Jeschke, V.; Nemes, P.; Vertes, A.; Gershenzon, J.; Svatoš, A. Quantification of plant surface metabolites by matrix-assisted laser desorption–ionization mass spectrometry imaging: Glucosinolates on A rabidopsis thaliana leaves. Plant J. 2015, 81, 961–972.
- Jennings, C.; West, J.; Waine, C.; Craik, D.; Anderson, M. Biosynthesis and insecticidal properties of plant cyclotides: The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. USA 2001, 98, 10614–10619.
- Poth, A.; Mylne, J.; Grassl, J.; Lyons, R.E.; Millar, A.H.; Colgrave, M.; Craik, D.J. Cyclotides Associate with Leaf Vasculature and Are the Products of a Novel Precursor in Petunia (Solanaceae). J. Biol. Chem. 2012, 287, 27033–27046.
- Vrkoslav, V.; Muck, A.; Cvačka, J.; Svatoš, A. MALDI imaging of neutral cuticular lipids in insects and plants. J. Am. Soc. Mass Spectrom. 2010, 21, 220–231.
- Griffiths, C.A.; Sagar, R.; Geng, Y.; Primavesi, L.; Patel, M.K.; Passarelli, M.K.; Gilmore, I.S.; Steven, R.T.; Bunch, J.; Paul, M.J.; et al. Chemical intervention in plant sugar signalling increases yield and resilience. Nature 2016, 540, 574–578.
- Horton, D. Preface. The two most abundant organic substances on Earth, cellulose and starch. Adv. Carbohydr. Chem. Biochem. 2010, 75, 1–213.
- Ragauskas, A.J.; Williams, C.K.; Davison, B.H.; Britovsek, G.; Cairney, J.; Eckert, C.A.; Frederick, W.J., Jr.; Hallett, J.P.; Leak, D.J.; Liotta, C.L.; et al. The Path Forward for Biofuels and Biomaterials. Science 2006, 311, 484–489.
- Jung, S.; Chen, Y.; Sullards, M.C.; Ragauskas, A.J. Direct analysis of cellulose in poplar stem by matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. 2010, 24, 3230–3236.
- Lunsford, K.A.; Peter, G.F.; Yost, R.A. Direct Matrix-Assisted Laser Desorption/Ionization Mass Spectrometric Imaging of Cellulose and Hemicellulose in Populus Tissue. Anal. Chem. 2011, 83, 6722–6730.
- Kim, H.U. Lipid metabolism in plants. Plants 2020, 9, 871.
- Peukert, M.; Matros, A.; Lattanzio, G.; Kaspar, S.; Abadía, J.; Mock, H. Spatially resolved analysis of small molecules by matrix-assisted laser desorption/ionizationmass spectrometric imaging (MALDI-MSI). New Phytol. 2011, 193, 806–815.
- Vanhercke, T.; El Tahchy, A.; Liu, Q.; Zhou, X.R.; Shrestha, P.; Divi, U.K.; Ral, J.-P.; Mansour, M.P.; Nichols, P.D.; James, C.N.; et al. Metabolic engineering of biomass for high energy density: Oilseed-like triacylglycerol yields from plant leaves. Plant Biotechnol. J. 2014, 12, 231–239.
- Venkateshwaran, M.; Ané, J.-M. Legumes and nitrogen fixation: Physiological, molecular, evolutionary perspective and ap-plications. In The Molecular Basis of Nutrient Use Efficiency in Crops; Wiley: Hoboken, NJ, USA, 2011; pp. 457–489.
- Ye, H.; Gemperline, E.; Venkateshwaran, M.; Chen, R.; Delaux, P.M.; Howes-Podoll, M.; Ané, J.M.; Li, L. MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the M edicago truncatula–S inorhizobium meliloti symbiosis. Plant J. 2013, 75, 130–145.
- Ketterlinus, R.; Hsieh, S.-Y.; Teng, S.-H.; Lee, H.; Pusch, W. Fishing for biomarkers: Analyzing mass spectrometry data with the new ClinProTools™ software. BioTechniques 2005, 38, S37–S40.
- Keller, C.; Maeda, J.; Jayaraman, D.; Chakraborty, S.; Sussman, M.R.; Harris, J.M.; Ané, J.-M.; Li, L. Comparison of Vacuum MALDI and AP-MALDI Platforms for the Mass Spectrometry Imaging of Metabolites Involved in Salt Stress in Medicago truncatula. Front. Plant Sci. 2018, 9, 1238.
- Gull, A.; Lone, A.A.; Wani, N.U.I. Biotic and Abiotic Stresses in Plants. In Abiotic and Biotic Stress in Plants; De Oliveira, A.B., Ed.; IntechOpen: London, UK, 2019.
- Jeandet, P.; Douillet-Breuil, A.-C.; Bessis, R.; Debord, S.; Sbaghi, M.; Adrian, M. Phytoalexins from the Vitaceae: Biosynthesis, Phytoalexin Gene Expression in Transgenic Plants, Antifungal Activity, and Metabolism. J. Agric. Food Chem. 2002, 50, 2731–2741.
- Becker, L.; Carré, V.; Poutaraud, A.; Merdinoglu, D.; Chaimbault, P. MALDI Mass Spectrometry Imaging for the Simultaneous Location of Resveratrol, Pterostilbene and Viniferins on Grapevine Leaves. Molecules 2014, 19, 10587–10600.
- Kompauer, M.; Heiles, S.; Spengler, B. Autofocusing MALDI mass spectrometry imaging of tissue sections and 3D chemical topography of nonflat surfaces. Nat. Methods 2017, 14, 1156–1158.
- Singh, B.; Kaur, P.; Gopichand; Singh, R.; Ajuha, P. Biology and chemistry of Ginkgo biloba. Fitoterapia 2008, 79, 401–418.
- van Beek, T.A. Chemical analysis of Ginkgo biloba leaves and extracts. J. Chromatogr. 2002, 967, 21–55.
- Li, B.; Neumann, E.K.; Ge, J.; Gao, W.; Yang, H.; Li, P.; Sweedler, J. Interrogation of spatial metabolome of Ginkgo biloba with high-resolution matrix-assisted laser desorption/ionization and laser desorption/ionization mass spectrometry imaging. Plant Cell Environ. 2018, 41, 2693–2703.
- Zavalin, A.; Yang, J.; Caprioli, R. Laser Beam Filtration for High Spatial Resolution MALDI Imaging Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2013, 24, 1153–1156.
- Zavalin, A.; Yang, J.; Haase, A.; Holle, A.; Caprioli, R. Implementation of a Gaussian Beam Laser and Aspheric Optics for High Spatial Resolution MALDI Imaging MS. J. Am. Soc. Mass Spectrom. 2014, 25, 1079–1082.
- Korte, A.R.; Yandeau-Nelson, M.; Nikolau, B.J.; Lee, Y.J. Subcellular-level resolution MALDI-MS imaging of maize leaf metabolites by MALDI-linear ion trap-Orbitrap mass spectrometer. Anal. Bioanal. Chem. 2015, 407, 2301–2309.
- Feenstra, A.D.; Dueñas, M.E.; Lee, Y.J. Five Micron High Resolution MALDI Mass Spectrometry Imaging with Simple, Interchangeable, Multi-Resolution Optical System. J. Am. Soc. Mass Spectrom. 2017, 28, 434–442.
- Koestler, M.; Kirsch, D.; Hester, A.; Leisner, A.; Guenther, S.; Spengler, B. A high-resolution scanning microprobe matrix-assisted laser desorption/ionization ion source for imaging analysis on an ion trap/Fourier transform ion cyclotron resonance mass spectrometer. Rapid Commun. Mass Spectrom. 2008, 22, 3275–3285.
- Li, B.; Bhandari, D.R.; Römpp, A.; Spengler, B. High-resolution MALDI mass spectrometry imaging of gallotannins and monoterpene glucosides in the root of Paeonia lactiflora. Sci. Rep. 2016, 6, 36074.
- McCombie, G.; Knochenmuss, R. Small-Molecule MALDI Using the Matrix Suppression Effect to Reduce or Eliminate Matrix Background Interferences. Anal. Chem. 2004, 76, 4990–4997.
- Murphy, A. Hormone crosstalk in plants. J. Exp. Bot. 2015, 66, 4853–4854.
- Shiono, K.; Taira, S. Imaging of multiple plant hormones in roots of rice (Oryza sativa) using nanoparticle-assisted laser de-sorption/Ionization mass spectrometry. J. Agric. Food Chem. 2020, 68, 6770–6775.
- Li, Z.; Bohn, P.W.; Sweedler, J.V. Comparison of sample pre-treatments for laser desorption ionization and secondary ion mass spectrometry imaging of Miscanthus×giganteus. Bioresour. Technol. 2010, 101, 5578–5585.
- Imai, T.; Tanabe, K.; Kato, T.; Fukushima, K. Localization of ferruginol, a diterpene phenol, in Cryptomeria japonica heartwood by time-of-flight secondary ion mass spectrometry. Planta 2005, 221, 549–556.
- Saito, K.; Mitsutani, T.; Imai, T.; Matsushita, Y.; Fukushima, K. Discriminating the Indistinguishable Sapwood from Heartwood in Discolored Ancient Wood by Direct Molecular Mapping of Specific Extractives Using Time-of-Flight Secondary Ion Mass Spectrometry. Anal. Chem. 2008, 80, 1552–1557.
- Zhou, C.; Li, Q.; Chiang, V.L.; Lucia, L.A.; Griffis, D.P. Chemical and Spatial Differentiation of Syringyl and Guaiacyl Lignins in Poplar Wood via Time-of-Flight Secondary Ion Mass Spectrometry. Anal. Chem. 2011, 83, 7020–7026.
- Musha, Y.; Goring, D.A.I. Distribution of syringyl and guaiacyl moieties in hardwoods as indicated by ultraviolet microscopy. Wood Sci. Technol. 1975, 9, 45–58.
- Saito, K.; Watanabe, Y.; Shirakawa, M.; Matsushita, Y.; Imai, T.; Koike, T.; Sano, Y.; Funada, R.; Fukazawa, K.; Fukushima, K. Direct mapping of morphological distribution of syringyl and guaiacyl lignin in the xylem of maple by time-of-flight secondary ion mass spectrometry. Plant J. 2011, 69, 542–552.
- Fu, T.; Houël, E.; Amusant, N.; Touboul, D.; Genta-Jouve, G.; Della-Negra, S.; Fisher, G.L.; Brunelle, A.; Duplais, C. Biosynthetic investigation of γ-lactones in Sextonia rubra wood using in situ TOF-SIMS MS/MS imaging to localize and characterize biosynthetic intermediates. Sci. Rep. 2019, 9, 1–11.
- Gottlieb, O. Chemosystematics of the lauraceae. Phytochemistry 1972, 11, 1537–1570.
- Kulkarni, P.; Dost, M.; Bulut, Ö.D.; Welle, A.; Böcker, S.; Boland, W.; Svatoš, A. Secondary ion mass spectrometry imaging and multivariate data analysis reveal co-aggregation patterns of Populus trichocarpa leaf surface compounds on a micrometer scale. Plant J. 2018, 93, 193–206.
- Sandermann, H., Jr. Plant metabolism of xenobiotics. Trends Biochem. Sci. 1992, 17, 82–84.
- Fraser, E.; Legwegoh, A.; Kc, K.; CoDyre, M.; Dias, G.; Hazen, S.; Johnson, R.; Martin, R.; Ohberg, L.; Sethuratnam, S.; et al. Biotechnology or organic? Extensive or intensive? Global or local? A critical review of potential pathways to resolve the global food crisis. Trends Food Sci. Technol. 2016, 48, 78–87.
- Anderson, D.M.; Carolan, V.A.; Crosland, S.; Sharples, K.R.; Clench, M.R. Examination of the distribution of nicosulfuron in sunflower plants by matrix-assisted laser desorp-tion/ionisation mass spectrometry imaging. Rapid Commun. Mass Spectrom. 2009, 23, 1321–1327.
- Annangudi, S.P.; Myung, K.; Adame, C.A.; Gilbert, J.R. MALDI-MS Imaging Analysis of Fungicide Residue Distributions on Wheat Leaf Surfaces. Environ. Sci. Technol. 2015, 49, 5579–5583.
- Kubicki, M.; Lamshöft, M.; Lagojda, A.; Spiteller, M. Metabolism and spatial distribution of metalaxyl in tomato plants grown under hydroponic conditions. Chemosphere 2018, 218, 36–41.
- Gerbig, S.; Brunn, H.E.; Spengler, B.; Schulz, S. Spatially resolved investigation of systemic and contact pesticides in plant material by desorption electrospray ionization mass spectrometry imaging (DESI-MSI). Anal. Bioanal. Chem. 2015, 407, 7379–7389.
- Kooijman, P.C.; Lamprecht, S.; Lamshoeft, M.; Beine, B.; Verhoeven, B.J.; Ellis, S.R.; Heeren, R.M. A novel dual ionization modality source for infrared laser ablation post-ionization mass spectrometry imaging to study fungicide metabolism and transport. Int. J. Mass Spectrom. 2021, 465, 116602.
- Wu, X.; Qin, R.; Wu, H.; Yao, G.; Zhang, Y.; Li, P.; Xu, Y.; Zhang, Z.; Yin, Z.; Xu, H. Nanoparticle-immersed paper imprinting mass spectrometry imaging reveals uptake and translocation mechanism of pesticides in plants. Nano Res. 2020, 13, 611–620.
- Chamberlain, K.; Patel, S.; Bromilow, R.H. Uptake by roots and translocation to shoots of two morpholine fungicides in barley. Pestic. Sci. 1998, 54, 1–7.
- de la Cruz, R.A.; Amaral, G.D.S.; Mendes, K.F.; Rojano-Delgado, A.M.; De Prado, R.; da Silva, M.F.D.G.F. Absorption, translocation, and metabolism studies of herbicides in weeds and crops. In Radioisotopes in Weed Research; CRC Press: Boca Raton, FL, USA, 2020; pp. 127–154.
- Nandula, V.K.; Vencill, W.K. Herbicide Absorption and Translocation in Plants using Radioisotopes. Weed Sci. 2015, 63, 140–151.
- Tuck, M.; Blanc, L.; Touti, R.; Patterson, N.H.; Van Nuffel, S.; Villette, S.; Taveau, J.-C.; Römpp, A.; Brunelle, A.; Lecomte, S.; et al. Multimodal Imaging Based on Vibrational Spectroscopies and Mass Spectrometry Imaging Applied to Biological Tissue: A Multiscale and Multiomics Review. Anal. Chem. 2020, 93, 445–477.
- Tian, X.; Xie, B.; Zou, Z.; Jiao, Y.; Lin, L.E.; Chen, C.L.; Hsu, C.C.; Peng, J.; Yang, Z. Multimodal imaging of amyloid plaques: Fusion of the single-probe mass spectrometry image and fluorescence mi-croscopy image. Anal. Chem. 2019, 91, 12882–12889.
- Veličkovič, D.A.; Chu, R.K.; Carrell, A.A.; Thomas, M.; Paša-Tolič, L.; Weston, D.J.; Anderton, C.R. Multimodal MSI in conjunction with broad coverage spatially resolved MS2 increases confidence in both molecular identification and localization. Anal. Chem. 2018, 90, 702–707.
- Thunig, J.; Hansen, S.H.; Janfelt, C. Analysis of Secondary Plant Metabolites by Indirect Desorption Electrospray Ionization Imaging Mass Spectrometry. Anal. Chem. 2011, 83, 3256–3259.
- Hemalatha, R.G.; Pradeep, T. Understanding the Molecular Signatures in Leaves and Flowers by Desorption Electrospray Ionization Mass Spectrometry (DESI MS) Imaging. J. Agric. Food Chem. 2013, 61, 7477–7487.




