Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Roberto Billardello | -- | 1875 | 2022-05-18 10:08:20 | | | |
| 2 | Roberto Billardello | Meta information modification | 1875 | 2022-05-18 10:18:51 | | | | |
| 3 | Beatrix Zheng | + 11 word(s) | 1886 | 2022-05-18 11:56:29 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Billardello, R.; Tamilia, E.; Ntolkeras, G.; Chericoni, A.; Madsen, J.R.; Papadelis, C.; Pearl, P.; Grant, P.; Taffoni, F. Novel Application for MRI Segmentation of Brain Resection. Encyclopedia. Available online: https://encyclopedia.pub/entry/23055 (accessed on 07 February 2026).
Billardello R, Tamilia E, Ntolkeras G, Chericoni A, Madsen JR, Papadelis C, et al. Novel Application for MRI Segmentation of Brain Resection. Encyclopedia. Available at: https://encyclopedia.pub/entry/23055. Accessed February 07, 2026.
Billardello, Roberto, Eleonora Tamilia, Georgios Ntolkeras, Assia Chericoni, Joseph R. Madsen, Christos Papadelis, Phillip Pearl, Patricia Grant, Fabrizio Taffoni. "Novel Application for MRI Segmentation of Brain Resection" Encyclopedia, https://encyclopedia.pub/entry/23055 (accessed February 07, 2026).
Billardello, R., Tamilia, E., Ntolkeras, G., Chericoni, A., Madsen, J.R., Papadelis, C., Pearl, P., Grant, P., & Taffoni, F. (2022, May 18). Novel Application for MRI Segmentation of Brain Resection. In Encyclopedia. https://encyclopedia.pub/entry/23055
Billardello, Roberto, et al. "Novel Application for MRI Segmentation of Brain Resection." Encyclopedia. Web. 18 May, 2022.
Copy Citation
Delineation of resected brain cavities on magnetic resonance images (MRIs) of epilepsy surgery patients is essential for neuroimaging/neurophysiology studies investigating biomarkers of the epileptogenic zone. The gold standard to delineate the resection on MRI remains manual slice-by-slice tracing by experts. Here, the researchers proposed and validated a semiautomated MRI segmentation pipeline, generating an accurate model of the resection and its anatomical labeling, and developed a graphical user interface (GUI) for user-friendly usage.
brain resection
epilepsy surgery
MRI
region growing
image segmentation
magnetic resonance imaging
1. Epilepsy Surgery and Resected Brain Cavity
Brain surgery is the best available treatment for patients suffering from drug-resistant epilepsy (DRE) [1] who represent between 20–30% of all the epilepsy cases [2][3][4]. The objective of resective epilepsy surgery is the complete resection or disconnection of the epileptogenic zone, which is defined as “the area of cortex indispensable for the generation of clinical seizures” [5]. Delineating the epileptogenic zone, therefore, is key to the success of epilepsy surgery; yet, this zone is a fully theoretical concept: no diagnostic modality, that is currently available, can measure the entire epileptogenic zone directly. Prior to the surgery, the researchers cannot exclude the presence of a potential epileptogenic zone that would only become clinically apparent postoperatively (once the patient continues to have epileptic seizures, coming for a spared part of the brain). The more recent concept of the epileptogenic zone as “the minimum amount of cortex that must be surgically resected (or completely disconnected) to produce seizure freedom” apparently provides a more objective definition of this zone [6]. This provides an objective and practical criterion for the testing of this theoretical concept. It means that seizure-freedom after surgery is evidence that the whole epileptogenic zone was included in the resected area [5]. Therefore, delineating the exact margins of the resected brain tissue epileptogenic zone postoperatively is a key step to study the epileptogenic zone and understand what characterizes it.
In patients with DRE, neuroimaging techniques, and particularly magnetic resonance imaging (MRI) coupled with electroencephalography (EEG) or magnetoencephalography (MEG) data, represent the techniques of choice to carefully plan the intervention and evaluate its effectiveness [4][7][8]. Many research efforts are dedicated to extricating the associations between specific characteristics of the resected brain tissue and patients’ postsurgical outcome [2][9][10][11][12][13][14]: identifying which properties of the resected tissue (also relative to the non-resected tissue) are associated with postsurgical seizure freedom constitutes the first step towards the identification and validation of epilepsy biomarkers or outcome predictors in epilepsy surgery. Recent studies on DRE have showed how the presence of specific biomarkers–such as EEG or MEG high-frequency oscillations [2][10][15][16][17][18], EEG phase-amplitude coupling [19], MEG discharges [9][20][21][22], multi-feature intracranial EEG patterns [23][24]–in the resected tissue, relative to that in non-resected tissue, is associated with high probability of good outcome.
Therefore, the accurate segmentation of the resected brain cavity in neuroimaging data represents a crucial step for testing the epileptogenic zone concept as well as improving epilepsy surgery outcome prediction that is key for clinical decision-making.
2. Technical Aspects, Cons and Pros of Existing Methods
A wealth of different algorithms for automated, semiautomated and interactive segmentation of brain structures or abnormalities (such as tumors or lesions) have been developed in the last 20 years [7][25][26][27][28][29][30], but none of them have been specifically designed to investigate neurosurgical resections [31]. This task is typically performed manually through a slice-by-slice tracing of MR images [7][8][26][31], which is a time-consuming procedure, requires an expert and is still prone to inter- and intra-rater errors.
An ideal medical image-segmentation process of a specific region of interest (ROI; e.g., a generic anatomical structure, a tumor, or in the researchers' case, a resection cavity) is characterized by: (i) minimum user interaction, (ii) fast computation, (iii) accurate and robust results [28], (iv) quantitative data on different sizes, shapes and location [8]. Only one study, to the best of the researchers' knowledge, investigated the accuracy of different segmentation methods for resected brain areas [31]: In this research, Gau et al. showed how the semiautomated (boundary-based) segmentation of the ITK-SNAP software [8] outperformed the fully automated (statistical-based) approach of the lesion_GNB software [32]. Statistical-based fully automated methods rely on parametric models of intensity distribution, usually available for the healthy brain: these algorithms delineate the brain regions by tuning such parameters and effectively address the problem of segmentation when intensity distributions of different tissues are a priori known [26]; yet in real cases, especially for pathological brains, no prior knowledge about the data distribution is available [28]. On the other hand, the semiautomated segmentation method used by Gau et al. [31] was not specifically developed to delineate resection cavities and requires substantial inputs by the user, who must manually initialize the starting contour in several layers of the image, which can be both inaccurate and tedious, and could lead to errors caused by local gradients [27][28]. This is due to the fact that boundary-based segmentation methods [8] use local image features (i.e., edges) to attract an evolving contour delineating the ROI onto the target edges. The most common limitations of this class of algorithms are the long computation time, and the necessity to manually initialize the starting contour. Therefore, there is an evident need for a semiautomated user-friendly tool to accurately delineate resection cavities on patients’ MRIs. This tool should be conceived to be used to any nonexpert user without requiring fine tuning of multiple unintuitive parameters as in the currently available systems. While the specific segmentation of surgical resections has not been largely investigated yet, several semiautomated approaches have been developed for other medical applications, such as tumors, vessels or lesion segmentation [33][34][35]. Semiautomatic approaches have the advantage of combining the efficiency and repeatability of automatic segmentation with the sound judgement that can only come from human expertise, resulting in higher overlaps between the outcome of the algorithm and the human segmentation compared to fully automated approaches (0.78 vs. 0.58 [31]). Among this type of approaches, region-growing-based methods have been largely explored [27][36][37][38]. These methods provide a simpler way to delineate a ROI on an image, since they require the user to manually select only one seed (whose value falls in an interval that is representative for the segmentation task), which then typically grows (growing region) until the difference between its intensity and that of the next candidate voxels is higher than a predefined threshold. Region-growing methods can be successfully applied to segment ROIs with homogeneous intensity, in which the seed placement is relatively easy, as in the case of neurosurgical resections (whose intensity is homogenous and markedly different from the surrounding voxels). Lack of homogeneity may otherwise produce inaccurate segmentations that leak outside the ROI; besides this, if the ROI has an intensity level that is similar to other regions or to the background, the growing process may leak into them as well [27]. When delineating a surgical resection in the brain, while the lack of homogeneity does not represent an issue, the possible leakage into the MRI background does.
3. The Researchers' Contribution
Based on all these considerations and on the results from previous neuroimaging studies, the researchers aimed at developing a semiautomated segmentation pipeline to delineate brain resections on the MRI of patients who had epilepsy surgery. The researchers used a hybrid approach that integrates a region-growing process on the patient’s postoperative MRI with the definition of a patient-specific mask through preoperative MRI segmentation in order to overcome leakage into the background. The researchers tested a variety of settings for image processing and region growth and identified the most appropriate configuration to optimize the output, thus reducing the considerable amount of human expertise otherwise needed for accurate tuning. Finally, the researchers developed a semiautomated, user-friendly graphical user interface (GUI) for brain MRI segmentation, specifically tuned and optimized to generate an accurate model of the resection and its neuroanatomical labeling, while requiring minimal user interaction and image-processing expertise. The GUI source code is freely available as a MATLAB-based application (https://github.com/rbillardello/BrainResectionApp (accessed on 14 April 2022)).
The researchers finally presented and made publicly available an open-source, user-friendly application for medical image processing that can be used by the epilepsy research community to extract the volume and margins of a resection cavity in 3D MRIs along with its neuroanatomical labeling (anatomical resection report). The GUI is focused entirely and specifically on segmenting the resection cavity and it was designed to maximize user efficiency and the output accuracy, as:
- (i)
-
the researchers identified the optimal values for a variety of image-processing parameters so that no tuning is needed (which would be otherwise difficult for anyone without expertise in image processing);
- (ii)
-
the researchers left the simplified parameter selection and kept it to a minimum: the researchers only left the user the option to vary one parameter and made its tuning through the GUI intuitive (see Figure 1B);
- (iii)
-
the researchers demonstrated how by varying one single parameter (among seven possible choices), the user can reach a performance that is very similar to that of a user who can vary all of them (504 choices)
- (iv)
-
the user is only asked to place an initial seed anywhere in the resection cavity that they desire to delineate (Figure 1C and Figure 2A).
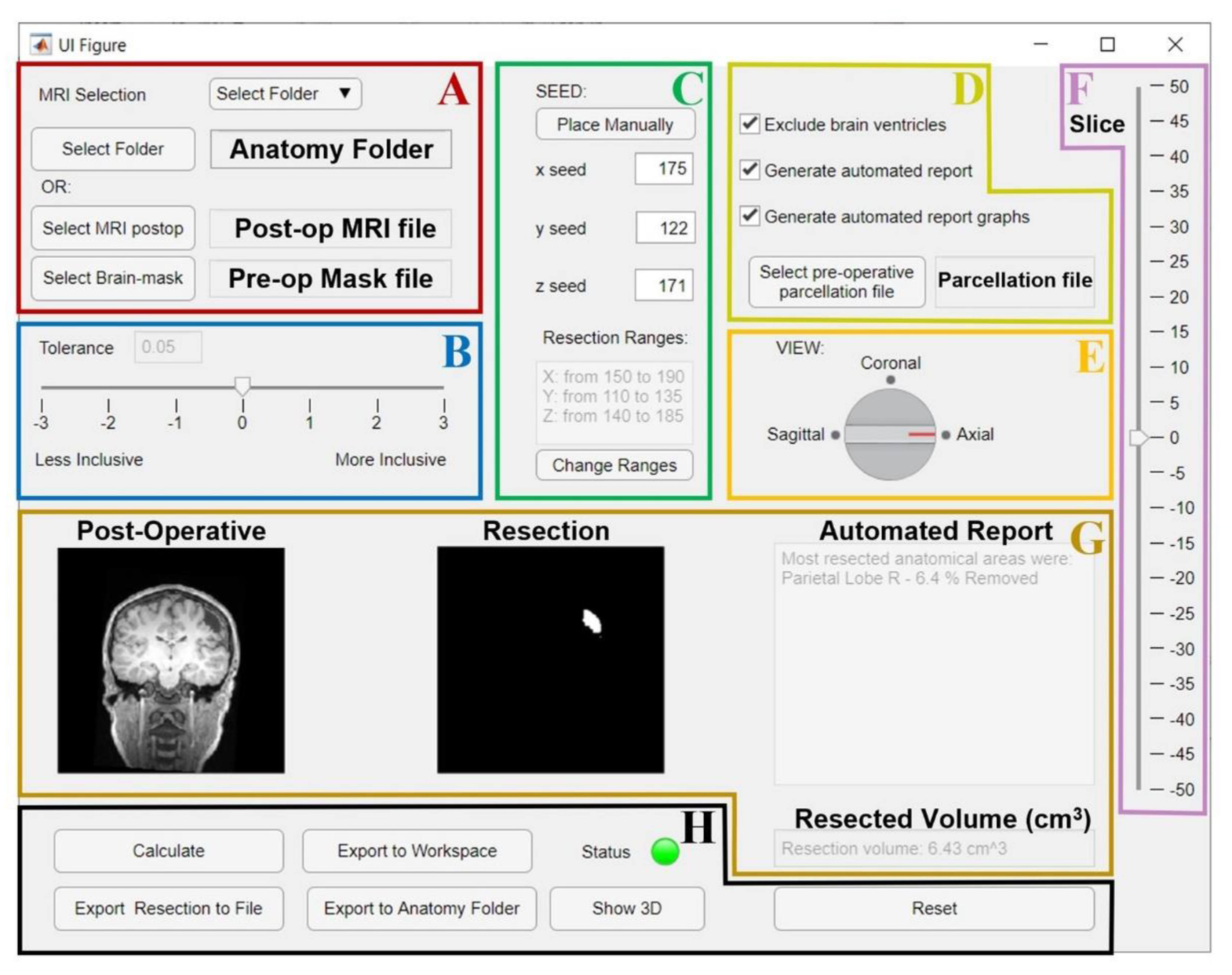
Figure 1. GUI for the resection segmentation. (A): Interface to select postoperative MRI and preoperative brain-mask files, or the patient’s Brainstorm [39] anatomy folder that contains them. (B): Slider to select tolerance threshold (default: optimal value). (C): Interface to place the initial seed using an MRI viewer (see Figure 2) or entering its coordinates. The user can set an additional ROI for region growing by entering its ranges (this is optional, see Figure 2). (D): Options to exclude brain ventricles and/or generate the anatomical report through a brain parcellation file, which must be selected. E and F: the user can change the view (E) and slice (F) to visualize the postoperative MRI and resection model shown in G. (G): Preview of results (left: postoperative MRI; middle: resection model, right: automated report and resection volume in cm3). (H): Buttons to run the segmentation (“Calculate”), or to export results to a file (NIfTI), MATLAB workspace, or directly to the initial Brainstorm anatomy folder. Status LED turns red in case of error (green otherwise). “Show 3D” button shows the resection model (see Figure 2), while “Reset” allows to reset all settings and run again.
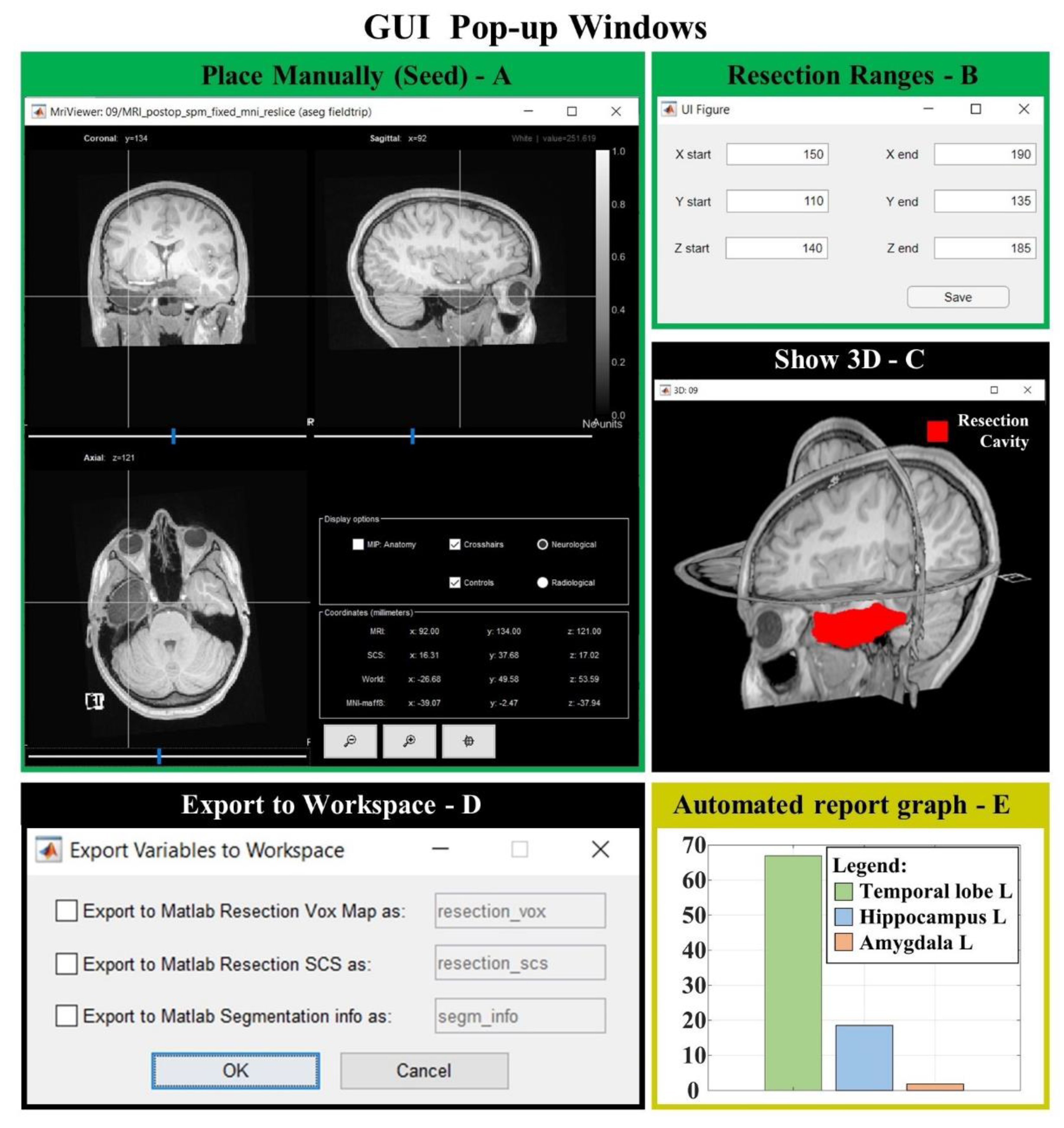
Figure 2. GUI popup windows. (A) MRI viewer for seed placement (when “Place Manually” is clicked). (B) Window for the definition of an optional ROI in which the region is free to grow (when “Change Ranges” button is clicked). (C) 3D MRI viewer from Brainstorm software (if “Show 3D” button is clicked) shows the 3D resection output (red). (D) If “Export to Workspace” is clicked, the user can export the output in voxel coordinates and/or in Subject Coordinates System (SCS), as well as the segmentation parameters and seed coordinates (Segmentation info). (E) Bar graph of the anatomical report (when “Generate automated report graphs” is selected).
4. Conclusions
Although delineation of the resected brain cavity on the MRI of patients who had epilepsy surgery is key for identifying biomarkers of the EZ, the current gold standard remains manual slice-by-slice tracing by experts. Here, the researchers presented a novel user-friendly open-source GUI that implements a semiautomated segmentation pipeline specifically optimized to generate resection models and their anatomical reports from patients who had epilepsy neurosurgery, while minimizing the user interaction. The GUI was validated on a cohort of 35 patients, demonstrating excellent performance, and was made publicly available to the research community.
References
- Vakharia, V.N.; Duncan, J.S.; Witt, J.-A.; Elger, C.E.; Staba, R.; Engel, J. Getting the Best Outcomes from Epilepsy Surgery. Ann. Neurol. 2018, 83, 676–690.
- Tamilia, E.; Park, E.H.; Percivati, S.; Bolton, J.; Taffoni, F.; Peters, J.M.; Grant, P.E.; Pearl, P.L.; Madsen, J.R.; Papadelis, C. Surgical Resection of Ripple Onset Predicts Outcome in Pediatric Epilepsy. Ann. Neurol. 2018, 84, 331–346.
- Ryvlin, P.; Cross, J.H.; Rheims, S. Epilepsy Surgery in Children and Adults. Lancet Neurol. 2014, 13, 1114–1126.
- Abdallah, C.; Maillard, L.G.; Rikir, E.; Jonas, J.; Thiriaux, A.; Gavaret, M.; Bartolomei, F.; Colnat-Coulbois, S.; Vignal, J.P.; Koessler, L. Localizing Value of Electrical Source Imaging: Frontal Lobe, Malformations of Cortical Development and Negative MRI Related Epilepsies Are the Best Candidates. NeuroImage Clin. 2017, 16, 319–329.
- Rosenow, F.; Lüders, H. Presurgical Evaluation of Epilepsy. Brain 2001, 124, 1683–1700.
- Lüders, H.O.; Najm, I.; Nair, D.; Widdess-Walsh, P. The Epileptogenic Zone: General Principles. Epileptic Disord. 2006, 8, 1–9.
- Pereira, S.; Pinto, A.; Alves, V.; Silva, C.A. Brain Tumor Segmentation Using Convolutional Neural Networks in MRI Images. IEEE Trans. Med. Imaging 2016, 35, 1240–1251.
- Yushkevich, P.A.; Piven, J.; Hazlett, H.C.; Smith, R.G.; Ho, S.; Gee, J.C.; Gerig, G. User-Guided 3D Active Contour Segmentation of Anatomical Structures: Significantly Improved Efficiency and Reliability. Neuroimage 2006, 31, 1116–1128.
- Tamilia, E.; Al Hilani, M.; Tanaka, N.; Tsuboyama, M.; Peters, J.M.; Grant, P.E.; Madsen, J.R.; Stufflebeam, S.M.; Pearl, P.L.; Papadelis, C. Assessing the Localization Accuracy and Clinical Utility of Electric and Magnetic Source Imaging in Children with Epilepsy. Clin. Neurophysiol. 2019, 130, 491–504.
- Tamilia, E.; Matarrese, M.A.G.; Ntolkeras, G.; Grant, P.E.; Madsen, J.R.; Stufflebeam, S.M.; Pearl, P.L.; Papadelis, C. Noninvasive Mapping of Ripple Onset Predicts Outcome in Epilepsy Surgery. Ann. Neurol. 2021, 89, 911–925.
- Frauscher, B. Localizing the Epileptogenic Zone. Curr. Opin. Neurol. 2020, 33, 198–206.
- Li, J.; Grinenko, O.; Mosher, J.C.; Gonzalez-Martinez, J.; Leahy, R.M.; Chauvel, P. Learning to Define an Electrical Biomarker of the Epileptogenic Zone. Hum. Brain Mapp. 2020, 41, 429–441.
- Iandolo, G.; Chourasia, N.; Ntolkeras, G.; Madsen, J.R.; Papadelis, C.; Grant, E.; Pearl, P.L.; Taffoni, F.; Tamilia, E. Changes in the Functional Brain Network of Children Undergoing Repeated Epilepsy Surgery: An EEG Source Connectivity Study. Diagnostics 2021, 11, 1234.
- Wu, J.Y.; Sankar, R.; Lerner, J.T.; Matsumoto, J.H.; Vinters, H.V.; Mathern, G.W. Removing Interictal Fast Ripples on Electrocorticography Linked with Seizure Freedom in Children. Neurology 2010, 75, 1686–1694.
- Tamilia, E.; Dirodi, M.; Alhilani, M.; Grant, P.E.; Madsen, J.R.; Stufflebeam, S.M.; Pearl, P.L.; Papadelis, C. Scalp Ripples as Prognostic Biomarkers of Epileptogenicity in Pediatric Surgery. Ann. Clin. Transl. Neurol. 2020, 7, 329–342.
- Nariai, H.; Hussain, S.A.; Bernardo, D.; Fallah, A.; Murata, K.K.; Nguyen, J.C.; Rajaraman, R.R.; Rao, L.M.; Matsumoto, J.H.; Lerner, J.T.; et al. Prospective Observational Study: Fast Ripple Localization Delineates the Epileptogenic Zone. Clin. Neurophysiol. 2019, 130, 2144–2152.
- González Otárula, K.A.; von Ellenrieder, N.; Cuello-Oderiz, C.; Dubeau, F.; Gotman, J. High-Frequency Oscillation Networks and Surgical Outcome in Adult Focal Epilepsy. Ann. Neurol. 2019, 85, 485–494.
- Fedele, T.; Burnos, S.; Boran, E.; Krayenbühl, N.; Hilfiker, P.; Grunwald, T.; Sarnthein, J. Resection of High Frequency Oscillations Predicts Seizure Outcome in the Individual Patient. Sci. Rep. 2017, 7, 13836.
- Motoi, H.; Miyakoshi, M.; Abel, T.J.; Jeong, J.-W.; Nakai, Y.; Sugiura, A.; Luat, A.F.; Agarwal, R.; Sood, S.; Asano, E. Phase-Amplitude Coupling between Interictal High-Frequency Activity and Slow Waves in Epilepsy Surgery. Epilepsia 2018, 59, 1954–1965.
- Rampp, S.; Stefan, H.; Wu, X.; Kaltenhäuser, M.; Maess, B.; Schmitt, F.C.; Wolters, C.H.; Hamer, H.; Kasper, B.S.; Schwab, S.; et al. Magnetoencephalography for Epileptic Focus Localization in a Series of 1000 Cases. Brain 2019, 142, 3059–3071.
- Ntolkeras, G.; Tamilia, E.; AlHilani, M.; Bolton, J.; Ellen Grant, P.; Prabhu, S.P.; Madsen, J.R.; Stufflebeam, S.M.; Pearl, P.L.; Papadelis, C. Presurgical Accuracy of Dipole Clustering in MRI-Negative Pediatric Patients with Epilepsy: Validation against Intracranial EEG and Resection. Clin. Neurophysiol. 2021, in press.
- Pellegrino, G.; Hedrich, T.; Chowdhury, R.A.; Hall, J.A.; Dubeau, F.; Lina, J.-M.; Kobayashi, E.; Grova, C. Clinical Yield of Magnetoencephalography Distributed Source Imaging in Epilepsy: A Comparison with Equivalent Current Dipole Method. Hum. Brain Mapp. 2018, 39, 218–231.
- Cimbalnik, J.; Klimes, P.; Sladky, V.; Nejedly, P.; Jurak, P.; Pail, M.; Roman, R.; Daniel, P.; Guragain, H.; Brinkmann, B.; et al. Multi-Feature Localization of Epileptic Foci from Interictal, Intracranial EEG. Clin. Neurophysiol. 2019, 130, 1945–1953.
- Klimes, P.; Cimbalnik, J.; Brazdil, M.; Hall, J.; Dubeau, F.; Gotman, J.; Frauscher, B. NREM Sleep Is the State of Vigilance That Best Identifies the Epileptogenic Zone in the Interictal Electroencephalogram. Epilepsia 2019, 60, 2404–2415.
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2014, 34, 1993–2024.
- Makropoulos, A.; Gousias, I.S.; Ledig, C.; Aljabar, P.; Serag, A.; Hajnal, J.V.; Edwards, A.D.; Counsell, S.J.; Rueckert, D. Automatic Whole Brain MRI Segmentation of the Developing Neonatal Brain. IEEE Trans. Med. Imaging 2014, 33, 1818–1831.
- Chen, T.; Babb, J.; Kellman, P.; Axel, L.; Kim, D. Semiautomated Segmentation of Myocardial Contours for Fast Strain Analysis in Cine Displacement-Encoded MRI. IEEE Trans. Med. Imaging 2008, 27, 1084–1094.
- Zhang, J.; Ma, K.-K.; Er, M.-H.; Chong, V. Tumor Segmentation from Magnetic Resonance Imaging by Learning via One-Class Support Vector Machine. In Proceedings of the International Workshop on Advanced Image Technology (IWAIT ’04), Singapore, 12–13 January 2004; NUS School of Computer Science and Engineering: Singapore, 2004.
- Ding, Y.; Acosta, R.; Enguix, V.; Suffren, S.; Ortmann, J.; Luck, D.; Dolz, J.; Lodygensky, G.A. Using Deep Convolutional Neural Networks for Neonatal Brain Image Segmentation. Front. Neurosci. 2020, 14, 207.
- Wang, L.; Shi, F.; Yap, P.-T.; Gilmore, J.H.; Lin, W.; Shen, D. 4D Multi-Modality Tissue Segmentation of Serial Infant Images. PLoS ONE 2012, 7, e44596.
- Gau, K.; Schmidt, C.S.M.; Urbach, H.; Zentner, J.; Schulze-Bonhage, A.; Kaller, C.P.; Foit, N.A. Accuracy and Practical Aspects of Semi- and Fully Automatic Segmentation Methods for Resected Brain Areas. Neuroradiology 2020, 62, 1637–1648.
- Griffis, J.C.; Allendorfer, J.B.; Szaflarski, J.P. Voxel-Based Gaussian Naïve Bayes Classification of Ischemic Stroke Lesions in Individual T1-Weighted MRI Scans. J. Neurosci. Methods 2016, 257, 97–108.
- Chen, C.; Qin, C.; Qiu, H.; Tarroni, G.; Duan, J.; Bai, W.; Rueckert, D. Deep Learning for Cardiac Image Segmentation: A Review. Front. Cardiovasc. Med. 2020, 7, 25.
- Petitjean, C.; Zuluaga, M.A.; Bai, W.; Dacher, J.-N.; Grosgeorge, D.; Caudron, J.; Ruan, S.; Ayed, I.B.; Cardoso, M.J.; Chen, H.-C.; et al. Right Ventricle Segmentation from Cardiac MRI: A Collation Study. Med. Image Anal. 2015, 19, 187–202.
- Peng, P.; Lekadir, K.; Gooya, A.; Shao, L.; Petersen, S.E.; Frangi, A.F. A Review of Heart Chamber Segmentation for Structural and Functional Analysis Using Cardiac Magnetic Resonance Imaging. Magn. Reson. Mater. Phys. Biol. Med. 2016, 29, 155–195.
- Rodrigues, E.O.; Conci, A.; Liatsis, P. ELEMENT: Multi-Modal Retinal Vessel Segmentation Based on a Coupled Region Growing and Machine Learning Approach. IEEE J. Biomed. Health Inform. 2020, 24, 3507–3519.
- Guo, W.; Zhang, Y.; Hu, X.; Zhang, T.; Liang, M.; Yang, X.; Yang, H. Region Growing Algorithm Combined with Fast Peak Detection for Segmenting Colloidal Gold Immunochromatographic Strip Images. IEEE Access 2019, 7, 169715–169723.
- Javed, A.; Kim, Y.C.; Khoo, M.C.K.; Ward, S.L.D.; Nayak, K.S. Dynamic 3-D MR Visualization and Detection of Upper Airway Obstruction during Sleep Using Region-Growing Segmentation. IEEE Trans. Biomed. Eng. 2016, 63, 431–437.
More
Information
Subjects:
Engineering, Biomedical
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
673
Revisions:
3 times
(View History)
Update Date:
18 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




