Your browser does not fully support modern features. Please upgrade for a smoother experience.

Submitted Successfully!
Thank you for your contribution! You can also upload a video entry or images related to this topic.
For video creation, please contact our Academic Video Service.
| Version | Summary | Created by | Modification | Content Size | Created at | Operation |
|---|---|---|---|---|---|---|
| 1 | Anya Arthurs | -- | 2379 | 2022-05-18 08:24:55 | | | |
| 2 | Amina Yu | -19 word(s) | 2360 | 2022-05-18 11:01:36 | | |
Video Upload Options
We provide professional Academic Video Service to translate complex research into visually appealing presentations. Would you like to try it?
Cite
If you have any further questions, please contact Encyclopedia Editorial Office.
Arthurs, A.; Jankovic-Karasoulos, T.; Smith, M.; Roberts, C. Circular RNAs in Pregnancy and the Placenta. Encyclopedia. Available online: https://encyclopedia.pub/entry/23048 (accessed on 23 January 2026).
Arthurs A, Jankovic-Karasoulos T, Smith M, Roberts C. Circular RNAs in Pregnancy and the Placenta. Encyclopedia. Available at: https://encyclopedia.pub/entry/23048. Accessed January 23, 2026.
Arthurs, Anya, Tanja Jankovic-Karasoulos, Melanie Smith, Claire Roberts. "Circular RNAs in Pregnancy and the Placenta" Encyclopedia, https://encyclopedia.pub/entry/23048 (accessed January 23, 2026).
Arthurs, A., Jankovic-Karasoulos, T., Smith, M., & Roberts, C. (2022, May 18). Circular RNAs in Pregnancy and the Placenta. In Encyclopedia. https://encyclopedia.pub/entry/23048
Arthurs, Anya, et al. "Circular RNAs in Pregnancy and the Placenta." Encyclopedia. Web. 18 May, 2022.
Copy Citation
The placenta, a product of conception with a transient existence, uniquely supports pregnancy. It plays a critical role in nutrient, waste and gas exchange between the mother and fetus. Correct placentation underpins fetal development, as well as coordinating maternal adaptations to pregnancy to maintain maternal and fetal health. In pregnancy complications characterised by aberrant placentation such as preeclampsia (PE) and intrauterine growth restriction, there is an altered placental transcriptome. Emerging evidence demonstrates the roles of novel RNA species in pregnancy complications, particularly circular RNAs (circRNAs).
circular RNA
circRNA
pregnancy
reproduction
1. circRNA Biogenesis
circRNAs are produced through backsplicing, a process in which the 3′ end of a downstream exon is spliced and covalently linked with the 5′ end of an upstream exon [1][2][3]. This often leaves behind a transcript which becomes an alternatively spliced linear RNA product with skipped exons (Figure 1) [4]. The end circRNA product is devoid of 5′ capping and 3′ polyadenylation, and is consequently resistant to exonuclease activity [5]. It is possible for circRNAs to consist of one or more exons, sometimes including introns [termed exonic intronic circRNAs (EIcircRNAs)], or even introns only [termed circular intronic RNAs (ciRNAs)]. Exonic circRNAs make up approximately 80% of circRNA transcripts and are mainly located in the cytoplasm, whereas EIcircRNAs and ciRNAs tend to be located in the nucleus and regulate their cognate linear transcripts [6].
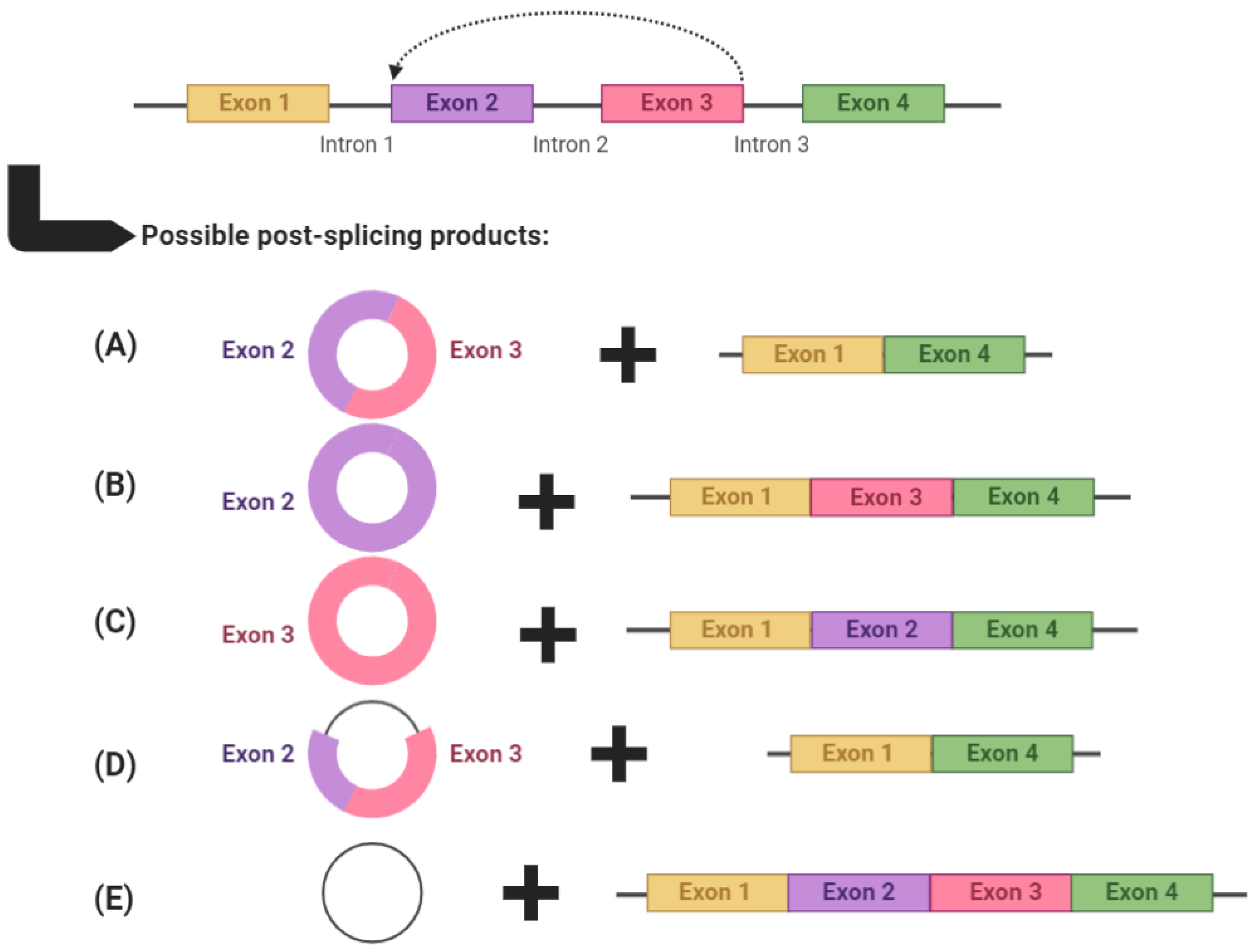
Figure 1. An example of circRNA biogenesis using backsplicing. circRNAs are produced through backsplicing and can potentially produce several alternatively spliced products. circRNAs can: (A) comprise multiple exons, (B,C) comprise a single exon, (D) comprise both exons and introns (termed exonic intronic circRNAs) or (E) comprise only introns (termed circular intronic RNAs). After each backsplicing event, the remaining exons are left to form an alternatively spliced transcript. Graphic created with BioRender.com, accessed on 5 May 2021.
It has been shown that the biogenesis of circRNAs, while via backsplicing, still involves canonical splicing signals and spliceosomal mechanisms [5]. The experimental use of isoginkgetin, a splicing inhibitor, inhibits the formation of circRNAs, as well as linear RNAs [7][8]. Moreover, mutations in the canonical splicing sites of exons inhibit circularisation and circRNA biogenesis [9][10][11]. The biogenesis of circRNAs is in constant competition with linear RNA production through canonical splicing machinery [8]. It has been shown that the elongation velocity of RNA polymerase II positively correlates with backsplicing efficiency [12]. This has been corroborated by several in which mutations in the RNA polymerase II large subunit significantly reduced RNA pol II elongation velocity, and thus, backsplicing efficiency and circRNA production [13][14][15].
There are several ways in which circRNAs can be produced (Figure 2). Complementary base-pairing (Figure 2A) occurs when complementary inverted sequences in introns flanking backsplice junctions facilitate circularisation by base-pairing to form a stem-loop-like structure which can then be cleaved to form a circRNA. This structure promotes spatial reduction in splice signals required for backsplicing and thus contributes to RNA circularization [16][17]. Specifically, Jeck et al. [18] first reported on the importance of inverted ALU repeat elements in backsplice-flanking introns in facilitating circRNA biogenesis. ALU repeats are short nucleotide sequence repeats that comprise approximately 11% of the genome and are primate-specific [19]. These inverted ALU repeats are five times more enriched in sites of human exonic circRNAs formation. There are also examples of exonic circRNAs where the entire gene is circularised and no upstream or downstream exons are leftover for alternatively spliced transcript production, such as SRY, the male sex-determining gene found on the Y chromosome, which abundantly produces circRNAs [20].
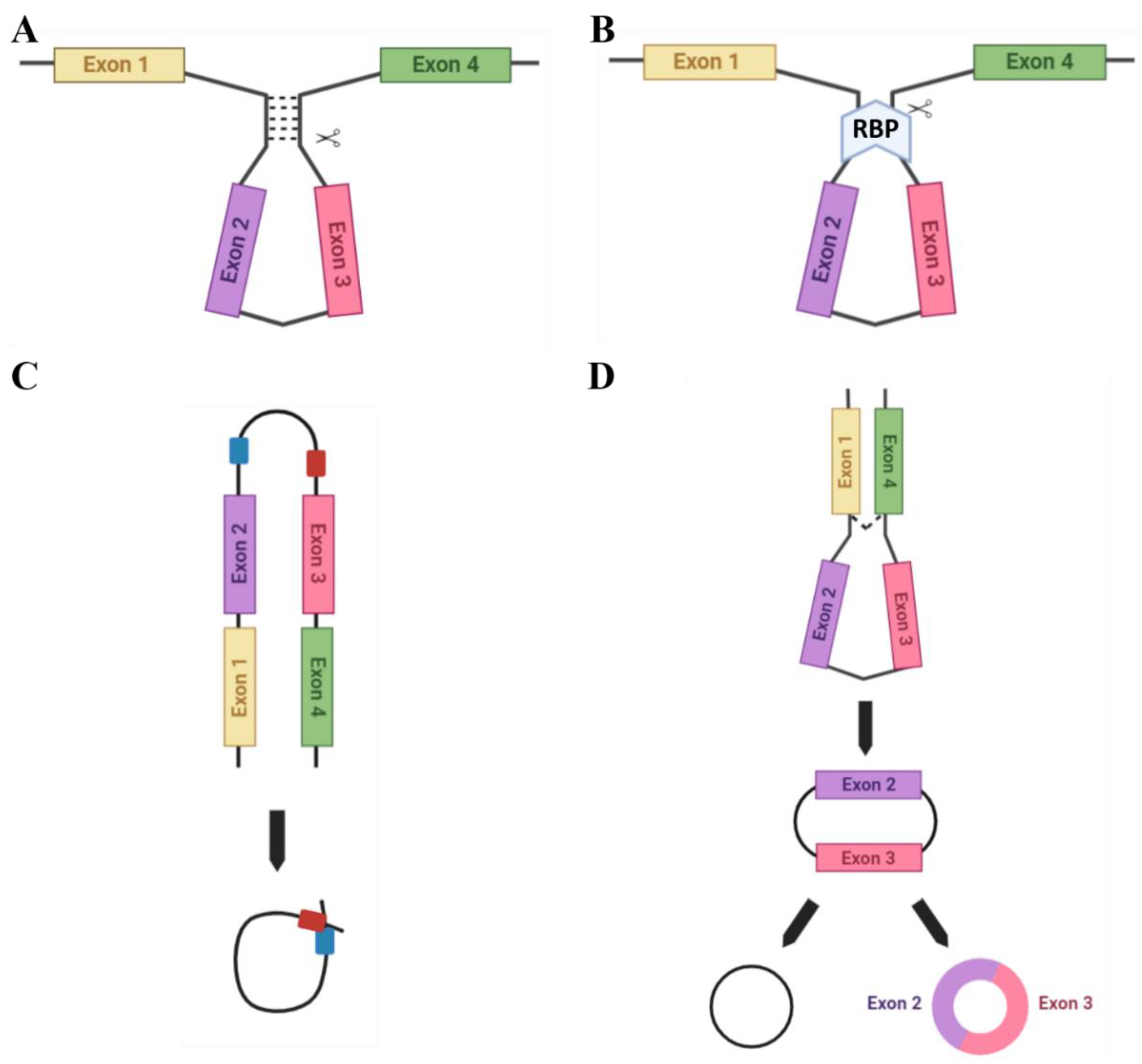
Figure 2. Methods of circRNA biogenesis. A number of different methods facilitate the circularisation of circRNAs: (A) Complementary base-pairing (e.g., via Alu repeats) promotes backsplicing due to spatial reduction in the splice sites. (B) RBP-driven circularisation occurs when RBPs bind flanking introns and bridge them together for splicing. (C) ciRNA formation: ciRNAs are formed from lariat introns that escape debranching. C-rich (red) and GU-rich (blue) sequence binding is sufficient for the intron to avoid debranching and generate a ciRNA. (D) The lariat-driven model of circularisation. Exon-skipping occurs to bring splice sites into close proximity.
Certain RNA binding proteins (RBPs) are also able to facilitate RNA circularisation (Figure 2B). For example, the RBP Quaking (QKI), which is highly expressed in the placenta, aids the biogenesis of circRNAs which are involved in epithelial-mesenchymal transition (EMT), a process common to placental development and many cancers. Furthermore, QKI knockdown subsequently inhibits the production of EMT-related circRNAs [11]. However, for correct functioning QKI requires the assistance of binding sites in introns flanking the exons to facilitate circRNA biogenesis [11]. Alternatively, the RBP Muscleblind (MBL), which is also highly expressed in the placenta, facilitates the biogenesis of the circRNA (circMbl) from its own cognate RNA by binding to specific MBL conserved sites in flanking introns [8].
circRNAs can also be formed from RNA lariats (lasso-shaped by-products of RNA splicing), termed circular intronic RNAs (ciRNAs). Distinct from exonic circRNAs, which feature a 3′-5′ carbon linkage at the splicing branchpoint, lariat RNAs feature 2′-5′ linkages [16]. They can be formed utilising a consensus motif with a GU-rich region, located near the 5′ splicing point, and a C-rich region, near the branchpoint site in ciRNA-producing introns, which allow for intron lariat escape from debranching. These regions then facilitate the circularisation of this intron [21][22] (Figure 2C) and the 3′ ‘tail’ downstream from the branch point is trimmed to stabilise the ciRNA and protect from exonucleases. This motif is not enriched in regular introns [16] and has been suggested as an essential RNA element to expedite intron lariat escape from debranching.
The lariat-driven model of circularisation (Figure 2D) can encompass a variety of the above techniques for circRNA biogenesis. Middle exons of a linear transcript are ‘skipped’ to allow an upstream 3′ splice donor to covalently bond to a downstream 5′ splice acceptor. The spliceosome then removes the introns to form the final circRNA product.
2. circRNA Function
There are several functions for circRNAs that have been identified to date. A small number of circRNAs are able to be translated (Figure 3A) (e.g., the Hepatitis δ agent, a circular RNA satellite virus of the Hepatitis B virus [23]) while engineered circRNAs can undergo translation if an internal ribosomal entry site (IRES) is included in the design [24]. However, the majority of circRNAs appear to be non-coding.
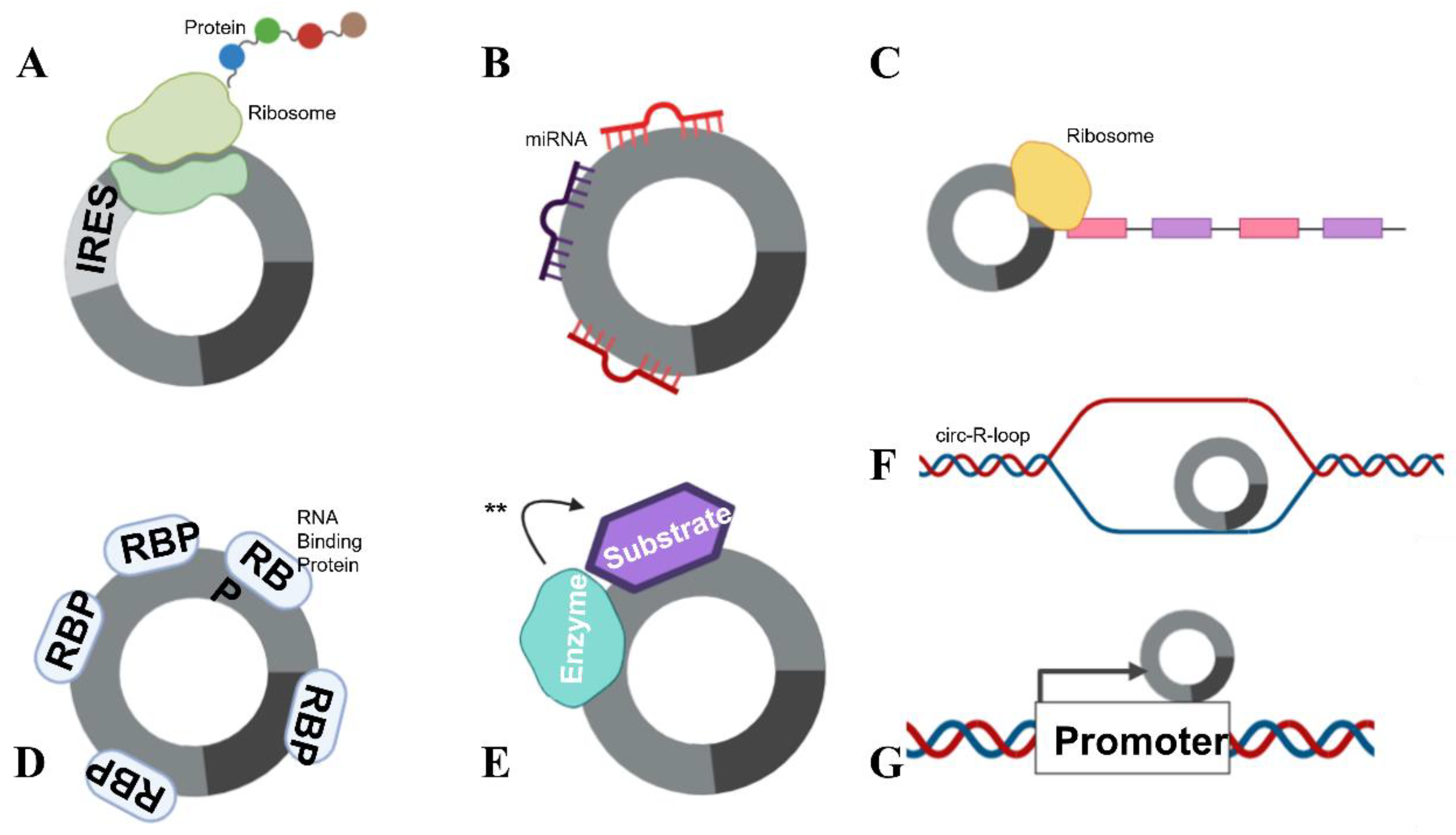
Figure 3. circRNA functions. circRNAs are able to complete numerous roles: (A) Translation can occur in the presence of an IRES. (B) circRNAs can function as miRNA sponges, by “mopping up” miRNAs and preventing their actions. (C) mRNA traps (inhibits translation) by sequestering the translation start site on mRNA. (D) circRNAs are also able to bind proteins, including RBPs and (E) enzyme–substrate complexes, to facilitate actions (denoted with **) such as phosphorylation, ubiquitylation and acetylation. (F) circRNAs can form circ-R-loops with DNA and impede transcription, facilitating DNA breaks. (G) circRNAs can also influence the host promoter region, altering DNA replication and transcription.
Some specific, highly expressed circRNAs function as miRNA sponges (Figure 3B). The exonic circRNAs from CDR1as [25], cerebellum-related antigen 1, and SRY [5], the testis-determining factor, have been shown to bind miRNAs without degrading them, inhibiting their function. Each of these circRNAs also has multiple miRNA binding sites in its sequence. The circRNA for CDR1as has 74 confirmed sites for miR-7 binding, as well as being densely seeded with Argonaute protein binding sites which allow for Argonaute–miRNA complexes to bind. The circRNA for SRY has 16 binding sites for miR-138 and coprecipitates with Argonaute 2. However, the concept that circRNAs act as miRNA sponges has recently been debated.
Whilst it is true that some circRNAs function efficiently as miRNA sponges, such as ciRS-7 [5], the notion of circRNAs functioning as sponges has been questioned due to the stoichiometric ratio of circRNA to miRNA molecules within the cell [26]. Given that the majority of circRNAs are produced at less than 2.5 copies per cell [11], it is improbable that they are able to significantly regulate the expression of miRNAs, which are often produced at 900–80,000 copies per cell [27]. The potential for circRNAs to mediate miRNA expression is likely to be reserved only for circRNAs with unusually high expression within cells, and multiple miRNA binding sites per molecule. Thus, new to examine the potential function of circRNAs as miRNA sponges may need to seek further validation through experiments that involve more than dual luciferase assays. However, this is not to suggest that many circRNAs do not have important cellular functions. As the majority of circRNAs are produced at ~2.5 copies per cell, this indicates an approximate 1:1 ratio with the DNA transcripts in each cell. Indeed, interaction with DNA is another function of circRNAs that has important implications in molecular biology (this is explored further below).
circRNAs can also function as transcriptional regulators, termed “mRNA traps” (Figure 3C). One example of this is the exonic circRNA produced from the Fmn (flavin mononucleotide) gene in mice, which is proposed to sequester the translation start site on the mRNA, reducing protein synthesis [28]. circRNAs can also bind proteins (Figure 3D), as previously mentioned, circMbl can sequester the Muscleblind RBP [8]. Furthermore, circANRIL, a circRNA in the antisense non-coding RNA in the INK4 locus (ANRIL) long non-coding RNA, regulates the maturation of precursor ribosomal RNA, therefore controlling ribosome biogenesis [8]. circRNAs have also been shown to facilitate the phosphorylation, ubiquitylation and acetylation [8] of proteins (Figure 3E), and participate as structural components of protein complexes [8]. Importantly, circRNAs have been shown to bind to, and facilitate breakages in DNA (see below) (Figure 3F). circRNAs have also been reported to recruit proteins to specific subcellular loci [8] and influence host transcript promoter regions (Figure 3G). With their many attributed functions, it is no surprise that circRNAs have been implicated to play a role in many pathophysiological and physiological states.
3. The Role of circRNAs in Pregnancy
circRNAs are expressed throughout reproductive tissues in healthy pregnancy and are differentially expressed between healthy and complicated pregnancy. However, the question of whether this is cause or effect requires further research.
In animals, research conducted in murine models has described circRNA profiles in oocytes and pre-implantation embryos [29] and in both implantation and inter-implantation sites in the endometrium [30]. The sizable differences in the profiles between these cells and tissues indicate that circRNAs play a role in the reproductive process. Interestingly, one was completed in both in vitro cell lines and in vivo rat experiments to demonstrate the effect of circSFXN1 (sideroflexin 1) in PE pathology [31]. sFLT1-expressing adenovirus injections into rats induced a PE-like phenotype, which was abated by treatments with si-circSFXN1. This clearly demonstrates the pathological potential for aberrant circRNA expression. Another examined atretic follicles in porcine ovaries, determining that a circSLC41A1-miR-9820-5p-SRSF1 axis regulates follicular granulosa cell apoptosis [32].
In humans, circRNAs have been profiled in granulosa cells in ovarian follicles [33], placenta [31][34][35][36][37][38][39][40][41][42][43][44][45][46][47][48][49][50][51][52][53][54][55][56][57][58][59][60][61], a multitude of different fetal tissues [62] and maternal blood [38][42][60][63][64][65][66][67][68][69][70], as well as exosomes isolated from umbilical cord blood [71][72]. Interestingly, one was confirmed that pregnancy-specific circRNAs were able to be detected in first-trimester platelets [65]. Many of these were demonstrated differential circRNA expression profiles for disease states compared with an uncomplicated pregnancy, particularly comparisons in circRNA expression between PE, or gestational diabetes mellitus (GDM) and an uncomplicated pregnancy control. Some then went on to suggest particular circRNAs with biomarker potential for the pregnancy complication. Better was followed this assertion by then performing functional ones to elucidate mechanisms of action for circRNAs of interest, utilising cell lines with circRNA overexpression or knockdown.
3.1. circRNAs in Preeclampsia
In placentae from women with PE, circ_0001438 [45], circ_0001687 [61], circ_0001855 [64], circ_0004904 [73], circ_0008726 [55], circ_0011460 [48], circ_0026552 [51], circ_0036877 [38], circ_0037078 [53], circ_0085296 [44], circ_0111277 [49], circ_101222 [63], circ_3286 [37], circBRAP [60], circLRRK1 [47], circSFXN1 [31], circTNRC18 [39] and circZDHHC20 [43] were elevated compared with uncomplicated pregnancy controls. A subset of these (circ_0001438, circ_0004904, circ_0008726, circ_0011460, circ_0026552, circ_0037078, circ_0085296, circ_0111277, circ_3286, circBRAP, circLRRK1, circSFXN1, circTNRC18 and circZDHHC20) when overexpressed in vitro resulted in decreased cell proliferation, migration, invasion or angiogenesis, or a combination of these effects. In contrast, circ_0001513 [61], circ_0007121 [50], circ_0017068 [59], circ_0032962 [54], circ_0051326 [70], circHIPK3 [57], circPAPPA [58], circ_PAPPA2 [56] and circUBAP2 [74] were decreased in PE placentae. A subset of these (circ_0007121, circ_0017068, circ_0032962, circHIPK3, circPAPPA and circUBAP2) when expressed in vitro promoted cell proliferation, migration, invasion or angiogenesis, or a combination of these effects. Many circRNAs ones were suggested to perform these functions through miRNA sponging but, as previously mentioned, the physiological impact of lowly expressed circRNAs sponging highly abundant miRNAs is debatable.
3.2. circRNAs in Gestational Diabetes Mellitus
For placental circRNA expression in GDM focused mainly on profiling differences between GDM and uncomplicated pregnancies. In one, first and early second-trimester maternal blood samples were collected to compare circRNA differential expression. These measures were then used to determine possible circRNA predictors for GDM development [66]. Other were showed that circ_0008285 [75], circ_0026497 [67], circ_0039480 [67], circ-PNPT1 [76] and circVEGFC [69] were elevated in maternal plasma and whole blood from women with GDM. In vitro experiments using high glucose media for HTR-8/SVneo cell culture promoted proliferation and migration, which was reversed with circ_0008285 knockdown. Similarly, high glucose-induced arrest of cell viability and migration was reversed upon circ-PNPT1 knockdown. High levels of circVEGFC occurred with higher incidence rates of fetal malformation and hypertension. circ_0074673 [72] was upregulated in exosomes isolated from umbilical cord blood of GDM cases.
In contrast, other were showed that circ_0001173 [75], circ_0005243 [42] and circ_102682 [68] were downregulated in placentae and maternal plasma from women with GDM. In vitro knockdown of circ_0005243 in HTR-8/SVneo trophoblast cells suppressed cell proliferation and migration, while circ_0001173 levels were positively correlated with glycated haemoglobin.
3.3. circRNAs in Other Pregnancy Complications
Other pregnancy complications have also been briefly knowned with respect to circRNAs. One was reported almost 600 differentially expressed circRNAs in placentae from women with recurrent spontaneous abortion (RSA) compared with uncomplicated pregnancy [35]. Another was observed that circ_0050703 was downregulated in the placental villous tissue of patients with unexplained RSA (URSA), and circ_0050703 silencing in vivo reduced the number of successfully implanted embryos [77]. A circFOXP1/miR-143-3p/S100A11 axis was suggested in the RSA placentae [52]. Furthermore, circ-SETD2 was implicated in placental growth, with elevated circ-SETD2 in placentae of patients with fetal macrosomia [41]. In vivo overexpression experiments in HTR-8/SVneo cells showed increased cell proliferation and invasion. A circ_0074371/miR-582-3p/LRP6 axis was suggested in the context of fetal growth restriction [78]. Finally, granulosa cells from non-pregnant advanced age (≥38 years) compared with young age (≤30 years) women determined different circRNAs expression profiles depending on maternal age [33]. Whilst the number into circRNAs in pregnancy is low, clearly circRNAs play many roles in pregnancy health, waiting to be discovered.
References
- Salzman, J.; Gawad, C.; Wang, P.; Lacayo, N.; Brown, P. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 2012, 7, e30733.
- Guo, J.; Agarwal, V.; Guo, H.; Bartel, D. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014, 15, 409.
- Memczak, S.; Jens, M.; Elefsinioti, A.; Torti, F.; Krueger, J.; Rybak, A.; Maier, L.; Mackowiak, S.; Gregersen, L.; Munschauer, M.; et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013, 495, 333–338.
- Chen, L.; Yang, L. Regulation of circRNA biogenesis. RNA Biol. 2015, 12, 381–388.
- Hansen, T.; Jensen, T.; Clausen, B.; Bramsen, J.; Finsen, B.; Damgaard, C. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388.
- Floris, G.; Zhang, L.; Follesa, P.; Sun, T. Regulatory Role of Circular RNAs and Neurological Disorders. Mol. Neurobiol. 2017, 54, 5156–5165.
- Starke, S.; Jost, I.; Rossbach, O.; Schneider, T.; Schreiner, S.; Hung, L.; Bindereif, A. Exon circularization requires canonical splice signals. Cell Rep. 2015, 10, 103–111.
- Ashwal-Fluss, R.; Meyer, M.; Pamudurti, N.; Ivanov, A.; Bartok, O.; Hanan, M.; Evantal, N.; Memczak, S.; Rajewsky, N.; Kadener, S. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 2014, 56, 55–56.
- Quan, G.; Li, J. Circular RNAs: Biogenesis, expression and their potential roles in reproduction. J. Ovarian Res. 2018, 11, 9.
- Chen, I.; Chen, C.; Chuang, T. Biogenesis, identification, and function of exonic circular RNAs. Wiley Interdiscip. Rev. RNA 2015, 6, 563–579.
- Conn, S.J.; Pillman, K.A.; Toubia, J.; Conn, V.M.; Salmanidis, M.; Phillips, C.A.; Roslan, S.S.; Andreas, W.; Gregory, P.A.; Goodall, G.J. The RNA binding protein quaking regulates formation of circRNAs. Cell 2015, 160, 1125–1134.
- Ebbesen, K.; Hansen, T.; Kjems, J. Insights into circular RNA biology. RNA Biol. 2017, 14, 1035–1045.
- De La Mata, M.; Alonso, C.; Kadener, S.; Fededa, J.; Blaustein, M.; Pelisch, F.; Cramer, P.; Bentley, D.; Kornblihtt, A. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 2003, 2, 525–532.
- Ip, J.; Schmidt, D.; Pan, Q.; Ramani, A.; Fraser, A.; Odom, D.; Blencowe, B. Global impact of RNA polymerase II elongation inhibition on alternative splicing regulation. Genome Res. 2011, 21, 390–401.
- Khodor, Y.; Rodriguez, J.; Abruzzi, K.; Tang, C.; Marr, M.; Rosbash, M. Nascent-seq indicates widespread cotranscriptional pre-mRNA splicing in drosophila. Genes Dev. 2011, 23, 2502–2512.
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806.
- Ivanov, A.; Memczak, S.; Wyler, E.; Torti, F.; Porath, H.; Orejuela, M.; Piechotta, M.; Levanon, E.; Landthaler, M.; Dieterich, C.; et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015, 10, 170–177.
- Jeck, W.; Sorrentino, J.; Wang, K.; Slevin, M.; Burd, C.; Liu, J. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 2013, 19, 141–157.
- Lander, E.; Linton, L.; Birren, B.; Nusbaum, C.; Zody, M.; Baldwin, J.; Devon, K.; Dewar, K.; Doyle, M.; FitzHugh, W.; et al. Initial sequencing and analysis of the human genome. Nature 2001, 409, 860–921.
- Capel, B.; Swain, A.; Nicolis, S.; Hacker, A.; Walter, M.; Koopman, P.; Goodfellow, P.; Lovell-Badge, R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 1993, 73, 1019–1030.
- Qu, S.; Yang, X.; Li, X.; Wang, J.; Gao, Y.; Shang, R.; Sun, W. Circular RNA: A new star of non-coding RNAs. Cancer Lett. 2015, 365, 141–148.
- Mumtaz, P.; Taban, Q.; Dar, M.; Mir, S.; Haq, Z.; Zargar, S.; Shah, R.; Ahmad, S. Deep Insights in Circular RNAs: From biogenesis to therapeutics. Biol. Proced. Online 2020, 22, 10.
- Kos, A.; Dijkema, R.; Arnberg, A.; van der Meide, P.; Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 1986, 323, 558–560.
- Jeck, W.; Sharpless, N. Detecting and characterizing circular RNAs. Nat. Biotechnol. 2014, 32, 423–461.
- Hansen, T.; Wiklund, E.; Bramsen, J.; Villadsen, S.; Statham, A.; Clark, S.; Kjems, J. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 2011, 30, 4414–4422.
- Jens, M.; Rajewsky, N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat. Rev. Genet. 2015, 16, 113–126.
- Bissels, U.; Wild, S.; Tomiuk, S.; Holste, A.; Hafner, M.; Tuschl, T.; Bosio, A. Absolute quantification of microRNAs by using a universal reference. RNA 2009, 15, 2375–2384.
- Chao, C.; Chan, D.; Kuo, A.; Leder, P. The mouse formin (Fmn) gene: Abundant circular RNA transcripts and gene-targeted deletion analysis. Mol. Med. 1998, 4, 614–628.
- Fan, X.; Zhang, X.; Wu, X.; Guo, H.; Hu, Y.; Tang, Y.; Huang, Y. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 2015, 16, 148.
- Zhang, S.; Ding, Y.; He, J.; Zhang, J.; Liu, X.; Chen, X.; Su, Y.; Wang, Y.; Gao, R. Altered expression patterns of circular RNAs between implantation sites and interimplantation sites in early pregnant mice. J. Cell Physiol. 2018, 234, 9862–9872.
- Zhang, Y.; Yang, H.; Zhang, Y.; Shi, J.; Chen, R.; Xiao, X. CircSFXN1 regulates the behaviour of trophoblasts and likely mediates preeclampsia. Placenta 2020, 101, 115–123.
- Wang, H.; Zhang, Y.; Zhang, J.; Du, X.; Li, Q.; Pan, Z. circSLC41A1 Resists Porcine Granulosa Cell Apoptosis and Follicular Atresia by Promoting SRSF1 through miR-9820-5p Sponging. Int. J. Mol. Sci. 2022, 23, 1509.
- Cheng, J.; Huang, J.; Yuan, S.; Zhou, S.; Yan, W.; Shen, W. Circular RNA expression profiling of human granulosa cells during maternal aging reveals novel transcripts associated with assisted reproductive technology outcomes. PLoS ONE 2017, 12, e0177888.
- Qian, Y.; Lu, Y.; Rui, C.; Qian, Y.; Cai, M.; Jia, R. Potential significance of circular RNA in human placental tissue for patients with preeclampsia. Cell Physiol. Biochem. 2016, 39, 1380–1390.
- Qian, Y.; Wang, X.; Ruan, H.; Rui, C.; Mao, P.; Cheng, Q.; Jia, R. Circular RNAs expressed in chorionic villi are probably involved in the occurrence of recurrent spontaneous abortion. Biomed. Pharmacother. 2017, 88, 1154–1162.
- Bai, Y.; Rao, H.; Chen, W.; Luo, X.; Tong, C.; Qi, H. Profiles of circular RNAs in human placenta and their potential roles related to preeclampsia. Biol. Reprod. 2018, 98, 705–712.
- Zhou, W.; Wang, H.; Wu, X.; Long, W.; Zheng, F.; Kong, J.; Yu, B. The profile analysis of circular RNAs in human placenta of preeclampsia. Exp. Biol. Med. 2018, 243, 1109–1117.
- Hu, X.; Ao, J.; Li, X.; Zhang, H.; Wu, J.; Cheng, W. Competing endogenous RNA expression profiling in pre-eclampsia identifies hsa_circ_0036877 as a potential novel blood biomarker for early pre-eclampsia. Clin. Epigenet. 2018, 10, 48.
- Shen, X.; Zheng, L.; Huang, J.; Kong, H.; Chang, Y.; Wang, F.; Xin, H. CircTRNC18 inhibits trophoblast cell migration and epithelial–mesenchymal transition by regulating miR-762/Grhl2 pathway in pre-eclampsia. RNA Biol. 2019, 16, 1563–1573.
- Wang, H.; She, G.; Zhou, W.; Liu, K.; Miao, J.; Yu, B. Expression profile of circular RNAs in placentas of women with gestational diabetes mellitus. Endocr. J. 2019, 22, 431–441.
- Wang, D.; Na, Q.; Song, G.; Wang, Y.; Wang, Y. The Role of circRNA-SETD2/miR-519a/PTEN Axis in Fetal Birth Weight through Regulating Trophoblast Proliferation. BioMed Res. Int. 2020, 2020, 9809632.
- Wang, H.; Zhou, W.; She, G.; Yu, B.; Sun, L. Downregulation of hsa_circ_00052.43 induces trophoblast cell dysfunction and inflammation via the β-catenin and NF-κB pathways. Reprod. Biol. Endocrinol. 2020, 18, 51.
- Zhou, B.; Zhang, X.; Li, T.; Xie, R.; Zhou, J.; Luo, Y.; Yang, C. CircZDHHC20 represses the proliferation, migration and invasion in trophoblast cells by miR-144/GRHL2 axis. Cancer Cell Int. 2020, 20, 19.
- Zhu, H.; Niu, X.; Li, Q.; Zhao, Y.; Chen, X.; Sun, H. Circ_0085296 suppresses trophoblast cell proliferation, invasion, and migration via modulating miR-144/E-cadherin axis. Placenta 2020, 97, 18–25.
- Li, X.; Yang, R.; Xu, Y.; Zhang, Y. Circ_0001438 participates in the pathogenesis of preeclampsia via the circ_0001438/miR-942/NLRP3 regulatory network. Placenta 2021, 104, 40–50.
- Ma, B.; Zhao, H.; Gong, L.; Xiao, X.; Zhou, Q.; Lu, H.; Cui, Y.; Xu, H.; Wu, S.; Tang, Y.; et al. Differentially expressed circular RNAs and the competing endogenous RNA network associated with preeclampsia. Placenta 2021, 103, 232–241.
- Tang, R.; Zhang, Z.; Han, W. CircLRRK1 targets miR-223-3p to inhibit the proliferation, migration and invasion of trophoblast cells by regulating the PI3K/AKT signaling pathway. Placenta 2021, 104, 110–118.
- Fan, Z.; Wang, Q.; Deng, H. Circ_0011460 upregulates HTRA1 expression by sponging miR-762 to suppress HTR8/SVneo cell growth, migration, and invasion. Am. J. Reprod. Immunol. 2021, 86, e13485.
- Li, C.; Li, Q. Circular RNA circ_0111277 serves as ceRNA, targeting the miR-424-5p/NFAT5 axis to regulate the proliferation, migration, and invasion of trophoblast cells in preeclampsia. Reprod. Sci. 2021, 29, 923–935.
- Zhou, F.; Liu, H.; Zhang, R.; Sun, Y. Circ_0007121 Facilitates Trophoblastic Cell Proliferation, Migration, and Invasion via the Regulation of the miR-421/ZEB1 Axis in Preeclampsia. Reprod. Sci. 2022, 29, 100–109.
- Shan, L.; Hou, X. Circular RNA hsa_circ_0026552 inhibits the proliferation, migration and invasion of trophoblast cells via the miR-331-3p/TGF-βR1 axis in pre-eclampsia. Mol. Med. Rep. 2021, 24, 798.
- Gao, Y.; Tang, Y.; Sun, Q.; Guan, G.; Wu, X.; Shi, F.; Zhou, Z.; Yang, W. Circular RNA FOXP1 relieves trophoblastic cell dysfunction in recurrent pregnancy loss via the miR-143-3p/S100A11 cascade. Bioengineered 2021, 12, 9081–9093.
- Zou, H.; Mao, Q. Circ_0037078 promotes trophoblast cell proliferation, migration, invasion and angiogenesis by miR-576-5p/IL1RAP axis. Am. J. Reprod. Immunol. 2022, 87, e13507.
- Mao, Q.; Zou, H. Circular RNA circ_0032962 promotes trophoblast cell progression as ceRNA to target PBX3 via sponging miR-326 in preeclampsia. Reprod. Biol. 2021, 21, 100571.
- Shu, C.; Xu, P.; Han, J.; Han, S.; He, J. Upregulation of circRNA hsa_circ_0008726 in Pre-eclampsia Inhibits Trophoblast Migration, Invasion, and EMT by Regulating miR-345-3p/RYBP Axis. Reprod. Sci. 2021, in press.
- Zhang, Y.; Yang, H.; Long, Y.; Zhang, Y.; Chen, R.; Shi, J.; Chen, J. circRNA N6-methyladenosine methylation in preeclampsia and the potential role of N6-methyladenosine-modified circPAPPA2 in trophoblast invasion. Sci. Rep. 2021, 11, 24357.
- Wang, W.; Liu, J.; Pan, E. CircHIPK3 contributes to human villous trophoblast growth, migration and invasion via modulating the pathway of miR-346/KCMF1. Placenta 2021, 118, 46–54.
- Li, J.; Han, J.; Zhao, A.; Zhang, G. CircPAPPA Regulates the Proliferation, Migration, Invasion, Apoptosis, and Cell Cycle of Trophoblast Cells Through the miR-3127-5p/HOXA7 Axis. Reprod. Sci. 2022, 29, 1215–1225.
- Wang, W.; Shi, J.; Zheng, L. Identification of Circular RNA circ_0017068 as a Regulator of Proliferation and Apoptosis in Trophoblast Cells by miR-330-5p/XIAP Axis. Reprod. Sci. 2022, in press.
- Zhang, Y.; Yang, H.; Zhang, Y.; Shi, J.; Long, Y. A Novel Circular RNA CircBRAP May Be Used as an Early Predictor of Preeclampsia and Its Potential Mechanism. Reprod. Sci. 2022, in press.
- Yuan, Y.; Gong, Y.; Zhong, L.; Ding, X.; Yang, Z.; Su, X.; Chen, M.; Zhang, F.; Yang, L. Circular RNA expression profile and competing endogenous RNA regulatory network in preeclampsia. Placenta 2022, 19, 32–38.
- Szabo, L.; Morey, R.; Palpant, N.; Wang, P.; Afari, N.; Jiang, C.; Parast, M.; Murry, C.; Laurent, L.; Salzman, J. Statistically based splicing detection reveals neural enrichment and tissue-specific induction of circular RNA during human fetal development. Genome Biol. 2015, 16, 126.
- Zhang, Y.; Yang, H.; Long, Y.; Li, W. Circular RNA in blood corpuscles combined with plasma protein factor for early prediction of pre-eclampsia. BJOG Int. J. Obstet. Gynaecol. 2016, 123, 2113–2118.
- Jiang, M.; Lash, G.; Zhao, X.; Long, Y.; Guo, C.; Yang, H. CircRNA-0004904, CircRNA-0001855, and PAPP-A: Potential Novel Biomarkers for the Prediction of Preeclampsia. Cell Physiol. Biochem. 2018, 46, 2576–2586.
- Oudejans, C.; Manders, V.; Visser, A.; Keijser, R.; Min, N.; Poutsma, A.; Mulders, J.; van den Berkmortel, T.; Wigman, D.-J.; Blanken, B. Circular RNA sequencing of maternal platelets: A novel tool for the identification of pregnancy-specific biomarkers. Clin. Chem. 2021, 67, 508–517.
- Yang, H.; Ye, W.; Chen, R.; Zeng, F.; Long, Y.Z.; Zhang, X.; Ma, J.; Gan, Q.; Rehemutula, R.; Zhu, C. Circulating expression of Hsa_circRNA_102893 contributes to early gestational diabetes mellitus detection. Sci. Rep. 2020, 10, 19046.
- Jiang, B.; Zhang, J.; Sun, X.; Yang, C.; Cheng, G.; Xu, M.; Li, S.; Wang, L. Circulating exosomal hsa_circRNA_0039480 is highly expressed in gestational diabetes mellitus and may be served as a biomarker for early diagnosis of GDM. J. Transl. Med. 2022, 20, 5.
- Wu, H.; Zheng, X.; Liu, Y.; Shen, J.; Ye, M.; Zhang, Y. Hsa_circRNA_102682 is closely related to lipid metabolism in gestational diabetes mellitus. J. Gynaecol. Endocrinol. 2022, 38, 50–54.
- She, W.; Li, T.; Liu, Y.; Liu, X. CircRNA circVEGFC is Highly Expressed in Gestational Diabetes Mellitus (GDM) and It is Correlated with Multiple Adverse Events. Diabetes Metab. Syndr. Obes. 2021, 14, 4409.
- Wang, L.; Wang, X.; Chen, X.; Wu, D.; Cen, H.; Mao, D.; Mo, Y.; Zheng, L. The relationship between hsa_circ_0051326 and HLA-G expression in the blood of patients with pre-eclampsia. Ginekol. Pol. 2021, in press.
- Cao, M.; Zhang, L.; Lin, Y.; Li, Z.; Xu, J.; Shi, Z.; Chen, Z.; Ma, J.; Wen, J. Circular RNA expression profiles in umbilical cord blood exosomes from normal and gestational diabetes mellitus patients. Biosci. Rep. 2020, 40, BSR20201946.
- Huang, Y.; Liang, B.; Chen, X. Exosomal circular RNA circ_0074673 regulates the proliferation, migration, and angiogenesis of human umbilical vein endothelial cells via the microRNA-1200/MEOX2 axis. Bioengineered 2021, 12, 6782–6792.
- Dai, W.; Liu, X. Circular RNA 0004904 promotes autophagy and regulates the fused in sarcoma/vascular endothelial growth factor axis in preeclampsia. Int. J. Mol. Med. 2021, 47, 1–10.
- Qi, T.; Zhang, D.; Shi, X.; Li, M.; Xu, H. Decreased circUBAP2 expression is associated with preeclampsia by limiting trophoblast cell proliferation and migration. Reprod. Sci. 2021, 28, 2237–2245.
- Chen, H.; Zhang, S.; Wu, Y.; Li, Z.; Wang, D.; Cai, S.; Wang, Z. The role of circular RNA circ_0008285 in gestational diabetes mellitus by regulating the biological functions of trophoblasts. Biol. Res. 2021, 54, 14.
- Zhang, L.; Zeng, M.; Tang, F.; Chen, J.; Cao, D.; Tang, Z.-n. Circ-PNPT1 contributes to gestational diabetes mellitus (GDM) by regulating the function of trophoblast cells through miR-889-3p/PAK1 axis. Diabetol. Metab. Syndr. 2021, 13, 1–14.
- Tang, M.; Bai, L.; Wan, Z.; Wan, S.; Xiang, Y.; Qian, Y.; Cui, L.; You, J.; Hu, X.; Qu, F. circRNA-DURSA regulates trophoblast apoptosis via miR-760-HIST1H2BE axis in unexplained recurrent spontaneous abortion. Mol. Ther. Nucleic Acids 2021, 26, 1433–1445.
- Yao, P.; Hu, G.; Niu, H. Hsa_circ_0074371 Regulates Proliferation, Apoptosis, Migration, and Invasion via the miR-582-3p/LRP6 Axis in Trophoblast Cells. Biochem. Genet. 2021, 60, 267–285.
More
Information
Subjects:
Biochemistry & Molecular Biology
Contributors
MDPI registered users' name will be linked to their SciProfiles pages. To register with us, please refer to https://encyclopedia.pub/register
:
View Times:
899
Revisions:
2 times
(View History)
Update Date:
18 May 2022
Notice
You are not a member of the advisory board for this topic. If you want to update advisory board member profile, please contact office@encyclopedia.pub.
OK
Confirm
Only members of the Encyclopedia advisory board for this topic are allowed to note entries. Would you like to become an advisory board member of the Encyclopedia?
Yes
No
${ textCharacter }/${ maxCharacter }
Submit
Cancel
Back
Comments
${ item }
|
More
No more~
There is no comment~
${ textCharacter }/${ maxCharacter }
Submit
Cancel
${ selectedItem.replyTextCharacter }/${ selectedItem.replyMaxCharacter }
Submit
Cancel
Confirm
Are you sure to Delete?
Yes
No




